We studied 1867 animals from 39 species including rodents, bats, primates and other wildlife species, from 6 provinces (Estuaire, Haut-Ogooué, Ogooué-Ivindo, Ogooué-Lolo, Ngounié and Woleu-Ntem) in Gabon. This study identified coronaviruses (CoVs) in 18 bats belonging to the species H. cf. ruber, H. gigas and M. inflatus from two caves located in the Ogooue-Ivindo province located northeast of Gabon. The nucleotides sequences obtained showed that these CoVs belonged to the Alphacoronavirus and Betacoronavirus genera. In a previous study, the identification of CoVs was already reported in H. cf. ruber in Gabon17. Taken together, these results indicated that genetically distinct coronaviruses co-circulate among bats in the Ogooue-Ivindo province. These findings supported that bats carry a great genetic diversity of CoVs and Woo et al.19 suggested that bats were ideal hosts for both alphacoronaviruses and betacoronaviruses and could play an important role in the ecology and evolution of coronaviruses. Although no coronavirus was identified in other animal species, alpha- and betacoronaviruses were already found naturally in various wild mammals such as civets5, buffalos20 and rodents21,22,23. In their studies in rodents from China, Lau et al.21 and Wang et al.22 analyzed 725 and 1465 rodents, respectively, whereas we analyzed only 494 rodents. The lack of detection of CoVs in our study could be linked to the small sample size, unlike the aforementioned studies. However, in their study, Ge et al.23 found 23 infected rodents out of the 177 studied. Unfortunately the rodents in our study come mainly from urban localities, rodent captures should also be considered in rural forest areas around caves inhabited by bats. Moreover, experimental infections have been described in non-human primates24,25 but no natural infection of CoV in NHP have yet been reported. For other wild animals, the small size of samples investigated by species could explain the lack of CoVs found in this study.
Alphacoronaviruses and betacoronaviruses were detected only in H. cf. ruber and H. gigas, 2 among the 5 species tested. These results from the Ogooue-Ivindo province suggest that these 2 species could be both natural reservoirs of and specific for the alpha- and the betacoronavirus, respectively. A close co-evolutionary association could be suspected between CoVs, particularly betacoronaviruses, and bats of this family26. Interestingly, in our study, we detected a specific Hipposideridae Betacoronavirus clustering with a Betacoronavirus previously detected in Thaïland in 200726. It was suggested that phylogenetic relationships between betacoronaviruses are mainly driven by the host phylogeny26. However, in a study conducted in Africa, Quan et al.27 suggested that there was no strict association between bat species and betacoronaviruses. Besides this study, previous studies on CoV host species in Asia have suggested that there was no strict association between bat species and alphacoronaviruses28,29. Indeed, many studies have described coronaviruses in a wide range of bat species30,31. Nevertheless, despite the report of alphacoronaviruses in numerous species of Miniopteridae and Vespertillionidae, Ar Gouilh et al.32 suggested a strict association between Alphacoronavirus EPI6 and Rhinolophidae, a familly usually associated with betacoronaviruses.
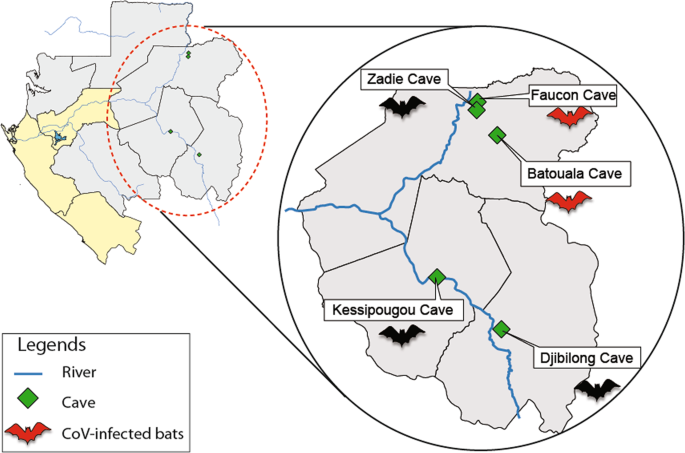
No CoVs were detected in chiropteran species sharing the same caves with the positive species (H. cf. ruber and H. gigas), suggesting a specificity of CoVs for these species or an absence of contact or interaction between some species. It was observed, for example, in the Faucon cave, that colonies of H. cf. ruber and H. gigas were found separately, in specific areas of the cave, the same with the other species (M. inflatus and C. afra), except for R. aegyptiacus which was not present in this cave. This spatial segregation could limit direct contact between species and represent a behavioral barrier to interspecific transmission, as previously described for lentiviruses and felids33,34 and more recently for bats and coronaviruses32.
A strain of Alphacoronavirus was identified in 7 H. cf. ruber nesting in the same cave (Faucon cave) and caught the same year (2009). Likewise, 2 other H. cf. ruber from the Batouala cave but caught in 2010 were infected by the same alphacoronavirus infecting the 7 H. cf. ruber from the Faucon cave. Corman et al.14 had already identified a closely related strain of Alphacoronavirus in 2 H. cf. ruber from the Faucon cave in 2009. Altogether, these findings first suggest the transmission of CoVs between individuals of the same species through close contacts probably increased by social and roosting behavior, large colony size and reduced space in these caves26; this strain of Alphacoronavirus could be enzootic in H. cf. ruber in this area of Gabon and roosts harboring a large number of bats nesting together promotes viral diffusion in the colony29,35, and second, local movements of bats individuals in the region imputable to seasonal/reproductive or opportunistic roosting behaviors, as the two caves being only 67 km apart. The migratory behavior of some bats provides an opportunity for pathogens to spread over long distances, as reported for flying foxes of the species Eidolon helvum able to migrate more than 1000 km36.
No individuals caught in 2011 and 2014 were found infected with CoVs. In 2011, only the guano of C. afra, H. cf. ruber, H. gigas, M. inflatus and R. aegyptiacus species was collected, unlike other years where organs were taken from bats. In this cave, the R. aegyptiacus colony lives in areas separated from other species including H. cf. ruber and H. gigas. The lack of detection in 2011 may be due to the poor quality of the samples having undergone several thawing. In 2014, only the R. aegyptiacus species was sampled in the Kessipoughou cave. In this cave, the R. aegyptiacus populations live in areas separated from other species including H. cf. ruber and H. gigas. However, Tong et al.37 and Lau et al.30 detected alphacoronaviruses in Rousettus bats in Kenya and China.
Alphacoronavirus strains detected in H. cf. ruber from the Faucon cave displayed a variable nucleotide identity of 91 to 93% with human coronavirus 229E. This virus causes a respiratory disease and is the cause of seasonal epidemics in humans38. In Ghana, Pfefferle et al.6 identified a strain of Alphacoronavirus in H. cf. ruber closely related to the human coronavirus 229E. Our finding argues that close relatives of the human coronavirus 229E exist in African bats14. The phylogenetic proximity of the CoV strains identified in this study with the human CoV-229E strain suggests a risk of emergence of this virus in humans in Africa. Furthermore, the 10GB0354 alphacoronavirus strain (MG963188), from H. gigas bats caught in 2010 in the Faucon cave, was phylogenetically close (93.8% nucleotide identity) to the Alpaca CoV strain detected in the Vicugna pacos species, commonly known as alpaca (JQ410000). Maganga et al.17 and Corman et al.14 already described two Gabonese CoV strains from hipposiderid bats caught in 2009 in the Faucon cave closely related to Alpaca CoV. Indeed, this coronavirus was described in 2007 during the epidemic of severe respiratory disorders associated with abortions in alpacas. It could be the direct progeny of its common ancestor with HCoV-229E, sharing with the latter a nucleotide identity of 92.2%39,40, on 440 bases of the nsp12 (polymerase). The phylogenetic link between these 2 CoVs could originate from an inter-species transmission, as a result of the opportunistic evolution of their common ancestor. As a result of genetic mutations and recombinations, the Alpaca CoV would have passed from bats to alpaca and vice versa. Phylogenetic analysis also revealed that two betacoronaviruses from 2 H. gigas, caught in 2013 in the Faucon cave, were phylogenetically close to a bat betacoronavirus named the Zaria bat coronavirus (ZCoV), detected in Nigeria in 2008 in a species of chiroptera genetically close to H. gigas, H. commersoni27, suggesting that phylogenetic proximity between host species may promote inter-species transmission41.
BLAST succeded in finding 2 contigs linked to coronavirus while no reads from this species could be detected by all EDGE’s taxonomy tools. To identify coronavirus reads having being assembled into those 2 contigs, further analyses using BLAST against NCBI RefSeq virus database were carried, allowing for the identification of 7 coronavirus reads. Since BLAST proved to be more sensitive, the same was done for the 2 other samples in order to verify that no coronavirus reads could have been missed. No more reads related to coronavirus were found. Moreover, this analysis overall showed that viral reads were extremely under-represented. One could argue that not finding the targeted virus might be caused by low DNA concentration in sequencing libraries. However, even in the sample where the highest concentration has been recorded, no coronavirus reads were found. Furthermore despite having achieved a satisfying quality sequencing, the biased amplification (majority species will be over-represented) and the low viral load explain why coronavirus was identified with such weak coverage.
Knowledge on the diversity of coronaviruses in bats and the factors promoting this diversity is important to prevent the risks of emergence in humans. Based on the analysis of data on life history traits of bats collected in the field, our findings suggest that the diversity of bat coronaviruses was associated with the seasonality and the site of capture, as well as the species of bats. It appeared that CoV infection in H. gigas correlated with the months of July and October, while CoV infection in H. cf. ruber was associated with the months of October and November.
In tropical ecosystems, some bat species seem to breed year-round, whereas others have annual or biannual birth peaks42. In Gabon, the parturitions in H. cf. ruber occur synchronously but some colonies give birth in March and others in October. In H. gigas from the Republic of Congo, a neighboring country, the young are born at the beginning of the rainy season. In Gabon, the months of October and November correspond to the beginning of the rainy season. A study demonstrated a high prevalence of CoVs in Chinese bats during the beginning of the rainy season43. The same finding was reported for the shedding of astroviruses in insectivorous bats in Asia (Borneo)42. Moreover, these months also correspond to the parturition season for H. gigas and H. cf. ruber. Indeed, lactating females of H. cf. ruber and juveniles of the same species were observed in October in northeastern Gabon. Regarding H. gigas, these observations are more difficult, Brosset and Saint Girons44 reported that the seasonal events of mating and parturition in H. gigas were confined in large caves where these large bats are entirely dependent to raise their young. According to Plowright et al.45, reproductive stress was an important driver of Hendra virus antibody prevalence. The prevalence of antibodies against the Hendra virus was higher in pregnant and lactating female of Pteropus scapulatus contrary to non-reproductive females. Different studies showed that parturition and lactation were important risk factors for CoVs shedding in insectivorous bats46,47. These two physiological states are energetically costly and thus lead to a depression of the immune system.
Moreover, our findings suggested that CoV infection was particularly associated with the Faucon cave. Of all the caves studied, the Faucon cave was the largest, with many cavities sheltering a diversity of bat species living in sympatry. This cave is regularly visited by villagers to hunt bats for human consumption. This hunting pressure could cause habitat disturbance and stress to these animals. Although Seltmann et al.42 found no association between habitat disturbance and the detection rate of CoVs in insectivorous bats, numerous studies in bats reported that pathogen prevalence was associated with habitat disturbance48. Many authors argued that habitat disturbance in some bats species may cause chronic stress and consequently immunosuppression42,49,50,51, increasing the susceptibility of animals to acquire and shed viruses.
In summary, the results of our study highlight a genetic diversity of CoVs in insectivorous bats in northeastern Gabon and show that the beginning of the rainy season and mainly the parturition season constitute a period of risk for CoV infection of insectivorous bats H. cf. ruber and H. gigas. These results also suggest an association between the disturbance of the bats’ habitat by human activities and the infection of these animals with these viruses. This study highlights a probable seasonality of the infection of insectivorous cave bats of the north-east of Gabon by CoVs. To conclude, a longitudinal monitoring should be carried out at different months of the year in order to determine the reproduction periods of the different bat species and therefore the periods favorable to viral shedding. The effect of the habitat disturbance on viral shedding should also be studied further. Bats harbor a large number of potentially zoonotic CoVs and pose a threat to public health for populations that are in contact with reservoir species of these viruses. The knowledge of CoVs ecology in bats is essential to prevent emergence in human populations.
Source: Ecology - nature.com


