In this study, intraspecific competition in C. maltaromaticum was mapped by performing high-throughput competition assays which investigated a total of 5 329 pairwise interactions within a collection of 73 strains. A high level of competition, at the intraspecific level, was recorded with approximatively 56% of the sender strains inhibiting at least one receiver strain and 82.2% of the receiver strains being inhibited by at least one sender strain. A previous study mapped interference competition at the interspecific level among 185 Vibrionaceae strains and revealed that 44% of the sender strains inhibited at least one other strain and 86% of the receiver strains were inhibited by at least one strain23. A more recent study mapped intergeneric interference competition within a collection of 37 gamma-proteobacteria belonging to different genera and showed that 81% of the sender strains antagonized at least one other strain and 81% of the receiver strains were antagonized by at least one strain12. This suggests that competition at the intraspecific level in C. maltaromaticum is relatively high because percentages of both antagonizing strains were higher within C. maltaromaticum population compared to the recorded percentages in the population of Vibrionaceae. Furthermore, when compared to intergeneric interference competition among gamma-proteobacteria, intraspecific competition in C. maltaromaticum was characterized by a slightly higher percentage of antagonized strains (82.2% of antagonized strains for C. maltaromaticum, and 81% of antagonized strains for gamma-proteobacteria). A high proportion of antagonism in C. maltaromaticum could be due to the fact that C. maltaromaticum strains were not collected from a same community. The majority of the strains were indeed isolated from different products (64 out of 73, see Supplementary Data S2). In Bacillus spp., the frequency of antagonism is indeed higher between bacteria isolated from different sites than bacteria isolated from the same site and was interpreted as a higher level of sympatric resistance15.
Besides the impact of geographic partitionning of the strains, the intraspecific phylogenetic scale and niche overlap could also explain the high frequency of antagonisms in C. maltaromaticum. Russel et al., 2017, by mapping interference competition within a collection of 67 bacterial species belonging to different phyla, demonstrated that inhibition was more frequent between phylogenetically related species characterized by a niche overlap. According to the authors, the main driver of competition seemed to be metabolic similarity that is the degree of niche overlap between the interacting microorganisms. The high level of competition obtained in C. maltaromaticum is in accordance with this theory since strains of this species have highly similar metabolism24. In line with this, numerous strains of C. maltaromaticum produce bacteriocins with anti-Listeria monocytogenes activity. This high frequency of anti-Listeria strains in this species is consistent with the fact that these species occupy similar niches: both species colonize food products such as dairy, meat and fish product, and both are psychrophilic bacteria.
C. maltaromaticum strains could be categorized into four different interactive profiles: PSR, SR, PR and R. About half of the strains (49.3%) exhibited a PSR profile while the PR profile was the rarest. Interestingly, modelling studies suggest that one important condition to meet in order to sustain microbial diversity despite interference competition is that all actors should exhibit a PSR profile10. However, this was not observed for all species. Prasad et al. 2011 were interested in delineating interacting strains of arctic bacteria according to their interactive profiles. In their study, they mapped antagonism among 139 strains isolated from five soil samples and they categorized strains of the four samples in which antagonism was observed. Seven different interactive profiles were identified, thus leading to a higher level of complexity compared to our study that lacks three profiles that were additionally observed within the collection of arctic bacteria: PS, P and S. Moreover, their results highly contrasted with the results obtained in this study, since the PSR profile was the less represented in all of the studied samples (the rarest in three samples and absent in one sample), whereas the R profile was the most common in two samples and the second most common in the other two samples. The reason explaining these differences is still unknown, future studies will be required to understand how microbial populations are shaped regarding these interacting profiles.
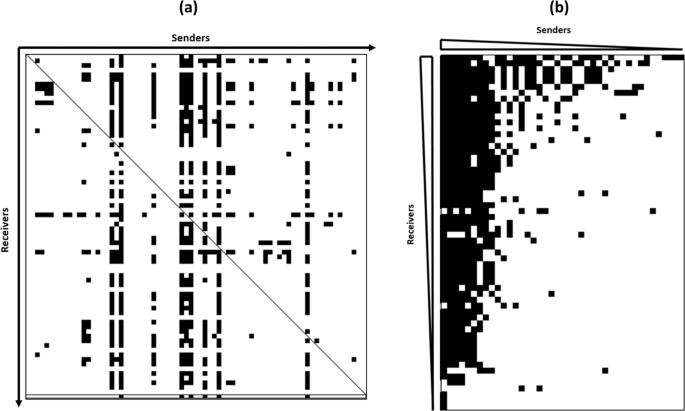
The competition network of C. maltaromaticum revealed a high variability of both the sender degrees characterizing the antagonistic activity spectra of sender strains and the receiver degrees characterizing the sensitivity spectra of receiver strains. Indeed, the sender degree is broad, ranging from 0 to 56 antagonized strains and peaks near extreme low values with 78% of the sender strains displaying low sender degrees (between 1 and 15). By contrast, the receiver degree is narrower than the sender degree, ranging from 0 to 27 antagonizing strains, and has an unimodal pattern. It was previously demonstrated for the competition network among 64 Streptomyces strains that the sender degree peaks near both extreme values, meaning that the majority of the sender strains are either slightly antagonistic or highly antagonistic13. This contrasts with the low frequency of highly antagonistic strains in C. maltaromaticum. This difference between Streptomyces and C. maltaromaticum could be explained by their different antagonistic potentialities; the genes encoding the antagonistic substances in Streptomyces genomes are more numerous and of a different type than those encoded by the species C. maltaromaticum and by lactic bacteria in general. Streptomyces produce various types of antimicrobial secondary metabolites including non ribosomal peptides, polyketones and bacteriocins25 while for C. maltaromaticum species, studies mainly characterized bacteriocins26,27. As for the distribution of receiver degrees, a similar narrow and unimodal pattern was observed in Streptomyces competition network, meaning that in C. maltaromaticum and Streptomyces, sensitive strains tended to have intermediate sensitivity levels, few were highly antagonized. Nevertheless, the sender-receiver asymmetry coefficient was very similar in both competition networks with a value of −0.38 in C. maltaromaticum and a value of −0.37 in Streptomyces. This implies that the outcome of competition is determined by the sender strains rather than the receiver strains. Surprisingly, the mapped competition among the 37 gamma-proteobacteria was also characterized by a sender-receiver asymmetry estimated by a value of – 0.31, very close to the previous ones12. This suggests that the sender determined asymmetry could be a common property of bacterial competition at many taxonomic levels, since it has been demonstrated for intergeneric competition12, interspecific competition13 and, by this present study, for intraspecific competition. More data should be generated from more bacterial competition network in order to test this hypothesis.
Sender-receiver asymmetry was found in C. maltaromaticum. In ecological terms, Vetsigian et al. (2011) stated that a sender-receiver asymmetry implies that the existence of an inhibitory interaction is more influenced by the sender which needs to produce an antagonistic substance than by the receiver which doesn’t need to resist to it. However, the analysis of C. maltaromaticum competition network revealed that antagonistic interactions are shaped by a strong pattern of nestedness (with a score of 6.59). This nestedness pattern indicates that a double determinism dictates the outcome of competition which depends on the the width of inhibition spectrum of sender strains and the width of sensitivity spectrum of the receiver strains. Similarly, Aguirre-von-Wobeser et al. 2014 were able to elucidate a nested structure of interference competition among 37 gamma-proteobacteria12 with a score of 0.872. The double determinism of inhibition implies that the competition is not randomly structured and the outcome of competition is related to the typology of both the sender and the receiver in terms of spectrum width.
The study of the reciprocity of the interactions in C. maltaromaticum revealed that reciprocal neutrality was the most frequent case (79.3% of total pairwise interactions). Approximately 20% percent of the remaining pairwise interactions were non-reciprocal inhibitory interactions, whereas only a minority were reciprocal inhibitory interactions (0.9% of total pairwise interactions). Such knowledge could be very useful for the selection of strains of this species in food production. Indeed, industrial food fermentation involves the use of industrial cultures. In the dairy industry for instance, milk is inoculated with cultures of selected microorganisms which act as major contributors of milk transformation to cheese or other fermented dairy products. These cultures often contain several different microorganisms. One major issue in this field of industrial culture is the assembly of these different microorganisms in order to get a synthetic community where each microorganism can cohabit. This requires reciprocal neutral interactions between the selected strains. Our results allowed us to identify strains exhibiting low potential of mutual inhibition in our experimental conditions. These strains are predicted to exhibit a high compatibility degree and could be co-cultivated without strong mutual competitive exclusion. These strains would be good candidates for the design of mixed starters.
This study revealed the extent and the structure of competition network in a highly genetically diversified LAB species. However, the study was conducted under laboratory conditions that poorly mime the natural conditions. It is therefore not known to what extent these competition interactions occur in ecosystems and to what extent the individual interaction profiles of strains could vary in a natural setting. Future studies are required to unravel the extent of interference competition in C. maltaromaticum in experimental models which take into account factors such as the the expression regulation of genes encoding antimicrobial substances, the stability and the diffusion of the secreted antimicrobial substances in the matrices.
Our study assessed competition that likely involves diffusible antimicrobial substances since the receiver strains were exposed to cell-free supernatant of sender strains. However, interference competition can also involve cell-cell contact28. It would also be interesting to study the extent of competition phenomena involving cell-cell contact and the structure of the resulting competition networks.
In addition, further studies of interspecific competition between diverse microbial species are needed in order to elucidate the role of the competition in microbial community assembly.
Source: Ecology - nature.com


