Diversity and composition of Antarctic penguin and tick RNA viromes
We characterised the transcriptomes of six libraries comprising ten individuals each, corresponding to three Antarctic penguin species in three locations and four tick libraries comprising a total of 20 ticks (Table 1 and Fig. 1). RNA sequencing of rRNA depleted libraries resulted in 42,382,642–55,930,902 reads assembled into 189,464–530,470 contigs for each of the penguin libraries. The tick libraries contained 51,498,136–55,930,902 reads assembled into 55,611–223,554 contigs (Table 1). There was a large range in the total viral abundance in both the penguin (0.07–0.7% total viral reads; 0–0.15% avian viral reads) and tick libraries (0.03–2.4%) (Table 1 and Fig. 2). In addition to likely avian viruses, the penguin libraries contained numerous reads matching insect, plant, or bacterial viruses and retroviruses (Figs. 2a and 3). Retroviruses were excluded from later analyses due to the challenges associated with differentiating exogenous from endogenous sequences.
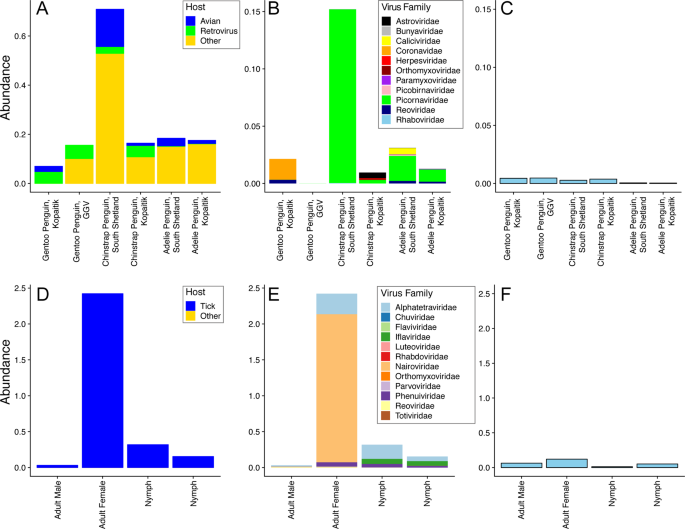
a Abundance of all viral reads found in penguin libraries. b Abundance and diversity of avian viruses in each of the penguin libraries. c Abundance of the host reference gene RSP13 in penguin libraries. d Abundance of all viral reads found in the tick libraries. e Abundance and diversity of viruses in each of the tick libraries. f Abundance of the host reference gene COX1 in the tick libraries.
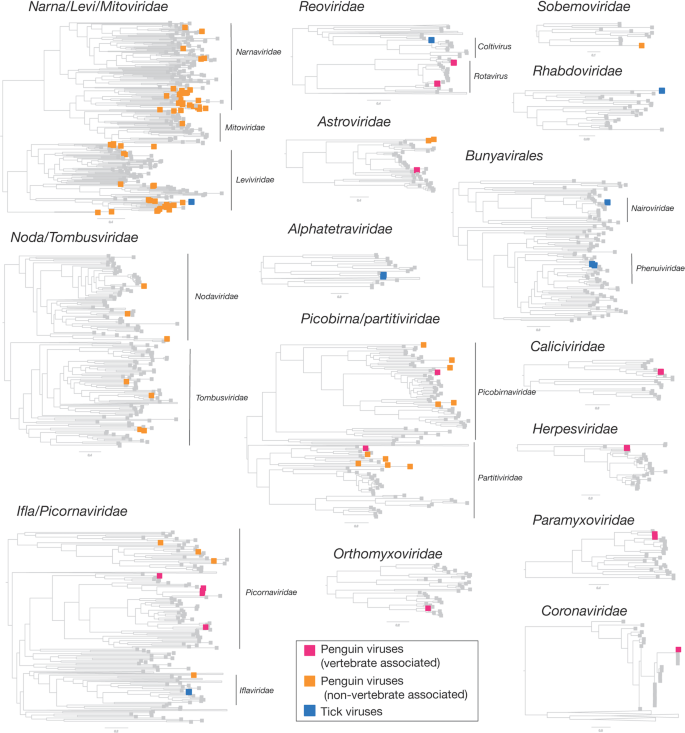
Viruses found in penguins were divided into two groups—those that infect birds and those that likely to other hosts and are coloured by magenta and orange boxes, respectively. Tick viruses revealed in this study are denoted by blue boxes. Grey boxes refer to reference viruses mined from RefSeq.
The abundance of RSP13, a stably expressed host reference gene in the avian colon [47], was similar across all penguin libraries, yet with lower abundance in the Adelie penguins (Fig. 2c). The abundance of COX1 in tick libraries was consistent with the body size of the ticks included in each library, with the highest abundance in the large adult female ticks and the lowest abundance in the first of two nymph libraries (Fig. 2d–f).
The abundance of avian viral reads was the highest in Chinstrap penguins sampled on Kopaitik Island (0.152% of total reads), and the lowest in Gentoo penguins sampled at the GGV Base for which no avian viral reads were detected. Because this colony is comprised solely of Gentoo penguins only this species was sampled [31]. Alpha diversity (the diversity within each library) was highest in the Adelie and Chinstrap penguins at both the viral family and genus levels, and was lower in Gentoo penguins, even when only considering RNA viromes from Kopaitik Island where all three penguin species were sampled (Figs. S1, S2). Hence, the reason we detected no viral reads at the most southern sampling site (the GGV Base) may be due to a combination of location and species choice (Gentoo penguins).
Although there was variation in virus composition among libraries, members of the Picornaviridae were the most abundant in the Chinstrap and Adelie penguin libraries, comprising 99, 32, 83, and 71% of all the avian viral reads in these four libraries. In marked contrast, the Picornaviridae comprised only 0.25% of avian viral reads from Gentoo penguins on Kopaitik Island (and no avian viruses were found in the Gentoo penguins from GGV). Beta diversity demonstrated connectivity in the RNA viromes from the Adelie penguins, driven by a number of shared viral species across the Kopaitik Island and King George Island sampling locations that are 130 km apart (Figs. 3 and S3).
Within the tick libraries, the greatest virus abundance was seen in the adult female ticks, while the lowest virus abundance was observed in the adult male ticks. Alpha diversity was similar across all libraries. Interestingly, while virus richness was highest in the adult female ticks, Shannon diversity was lower than the other libraries (Fig. S4), although without replicates it is not possible to draw clear conclusions. Given the high virus richness in female ticks, it is not surprising that the largest number of viral species were also described in this library. The tick libraries were also highly connected, with 5/8 species shared among them, although the beta diversity calculations are confounded because of limited sample size (Fig. S3).
Substantial RNA virus diversity in Antarctic penguins and their ticks
Overall, 22 viral families, in addition to four viruses that fell outside well defined viral families but clustered with other unclassified “picorna-like” viruses (Treshnikov virus, Luncke virus, Dralkin virus, and Tolstivok virus), were identified in the penguin and tick libraries (Fig. 3). Of these, the likely bird associated viruses were members of the Astroviridae, Caliciviridae, Coronaviridae, Herpesviridae, Orthomyxoviridae, Paramyxoviridae, Picorbirnaviridae, Picornaviridae, and Reoviridae (Figs. 2b and 3) (see below). Ten of the 13 avian associated viruses identified in the penguins likely represent novel avian viral species (Table S2 and Fig. 4), and two virus species (Avian avulavirus 17 and Shirase virus) were shared among Adelie penguins from different locations. There was no virus sharing among species at individual locations (i.e., no viruses were shared across penguin species at either Kopaitik Island or on King George Island) (Fig. 4), although this is likely because species were sampled in different years. All viruses in the tick libraries, with the exception of Taggert virus, represented novel virus species with amino acid sequence similarity to reference virus sequences from 31 to 81%. Five of the nine virus species described in ticks were shared across libraries, which is unsurprising given that the ticks were collected from the same population. Notably, the nymph libraries contained a higher percentage of viral RNA than both the large nymphs and adult male ticks.
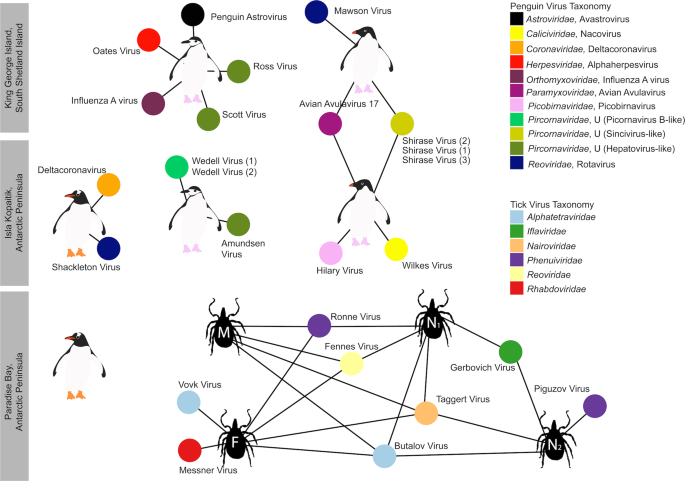
Each library is represented as a central node, with a pictogram of the species, surrounded by each viral species. Line lengths do not correspond to any variable. Where two libraries share a virus species, the networks between the two libraries are linked. Virus colour corresponds to virus taxonomy. Viruses identified in penguin libraries that are unlikely to be bird associated are not shown. A list of viruses from each library is presented in Table S2, and phylogenetic trees for each virus family can be found in Figs. 5–7 and S5–S13.
Strikingly, we identified 82 divergent novel virus species in the penguin libraries that clustered phylogenetically within 11 defined families, as well as three viruses that clustered with a group of unclassified viruses. These unclassified viruses are likely associated with penguin diet or their microbiome: fish, invertebrates, plants, fungi and bacteria (Fig. 2a, Table S3, Fig. 3). The largest diversity was found in the “Narna-Levi”, “Noda-Tombus” and “Picorbirna-Pariti” viral groups [36]. A number of viruses were highly divergent, including clusters of novel viruses that fell within the Narnaviridae and Leviviridae (Table S3 and Fig. 3). Overall, 56 different species of Narna-Levi viruses were identified in Adelie penguins, comprising approximately half of the Narna-like viruses and 21/25 of the Leviviridae: these were likely associated with bird diet or microbiome. All invertebrate associated Picobirnaviridae were found in Chinstrap penguin libraries, while a single picobirnavirus identified in an Adelie penguin library was most closely related to other bird associated viruses (see below). As these 82 viruses are unlikely to be associated with penguins or their ticks, they are not described further.
Novel avian viruses
The novel Wilkes virus was identified in an Adelie penguin on Kopaitik Island, and belongs to the genus Nacovirus (Caliciviridae)—a group dominated by avian viruses (Fig. S5). This virus is closely related to Goose calicivirus and caliciviruses sampled from waterbirds in Australia (i.e., Red-necked Avocet and Pink-eared Duck) [32, 48]. All the picornaviruses identified in this study likely belong to novel or unassigned genera (Fig. S6). Three different variants of Shirase virus were identified in Adelie penguins, two from King George Island and one from Kopaitik Island. Interestingly, Shirase virus falls as a sister lineage to viruses of the genus Gallivirus. Similarly, two variants of Wedell virus were identified in Chinstrap penguins. Wedell virus falls in an unassigned lineage of picornaviruses different than avian viruses identified in metagenomic studies [32, 48]. Three additional picornaviruses were identified in Chinstrap penguins—Ross virus, Scott virus, and Amundsen virus—that fall basal to members of the genus Tremovirus.
Rotaviruses were identified in both Gentoo (Shackleton virus) and Adelie (Mawson virus) penguins. Shackleton virus falls as an outgroup to a clade of rotaviruses recently described in wild birds, which are themselves divergent from rotavirus G virus, while Mawson virus is a sister group to rotavirus D (Fig. 5a). Hilary virus, a picobirnavirus, was found in a clade that contains both avian and mammalian hosts. Interestingly, this virus is most closely related to a human picorbirnavirus, albeit with low amino acid similarity and long branch lengths (Fig. S7). Although there is uncertainty as to whether these viruses are bacterial rather than vertebrate associated [49], they are retained here for comparative purposes.
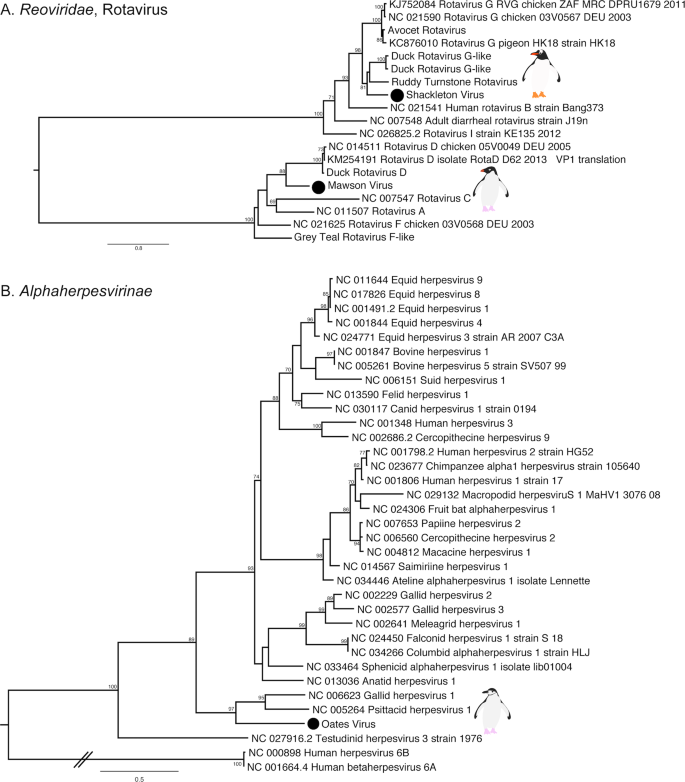
a Phylogenetic tree of the VP1, containing the RdRp, of rotaviruses. The tree is mid-point rooted for clarity only. b Phylogeny of the concatenated major capsid gene and glycoprotein B gene, the only genes recovered, of the Alphaherpesvirinae. Two betaherpesviruses were used as outgroup to root the tree. The viruses identified in this study are denoted with a filled circle and in bold. Bootstrap values >70% are shown for key nodes. The scale bar represents the number of amino acid substitutions per site.
Finally, although most of the novel viruses documented here had RNA genomes, we also identified a novel alphaherpesvirus, Oates virus, that falls as a sister group to Gallid and Psittacid hepervirus 1. Notably, this virus was distantly related to an alphaherpesvirus previously described in penguins (Sphencid alphaherpesvirus) (Fig. 5b).
Avian RNA viruses previously detected in penguins
Previous studies of Antarctic penguins have detected avian IAV and avian avulaviruses [15, 16, 21]. Similarly, we detected an H5N5 IAV in Chinstrap penguins identical in sequence to that reported previously. This is not surprising as the virus described in Hurt et al. [15] was isolated in the same set of samples (Fig. S8). In addition, we identified Avian avulavirus 17 (AAvV-17) in Adelie penguins from both sampling locations (Figs. 6a and S9). This virus was previously isolated in Adelie penguins in 2013 [21] and Gentoo penguins in 2014–2016 [50]. Analysis of the F gene of AAvV-17 indicates that the virus detected in Adelie penguins on both King George Island and Kopaitik Island was more closely related to that from Gentoo penguins sampled between 2014 and 2016 [50] than to the Adelie penguins sampled in 2013 [21] (Fig. 6a). Although AAvV-17 was detected in penguins sampled at two locations (Kopaitik Island and King George Island) only 2 weeks apart, they shared only 98.6% identity. Blastx results also indicated the presence of Avian avulavirus 2, although we were unable to assemble the virus genome.
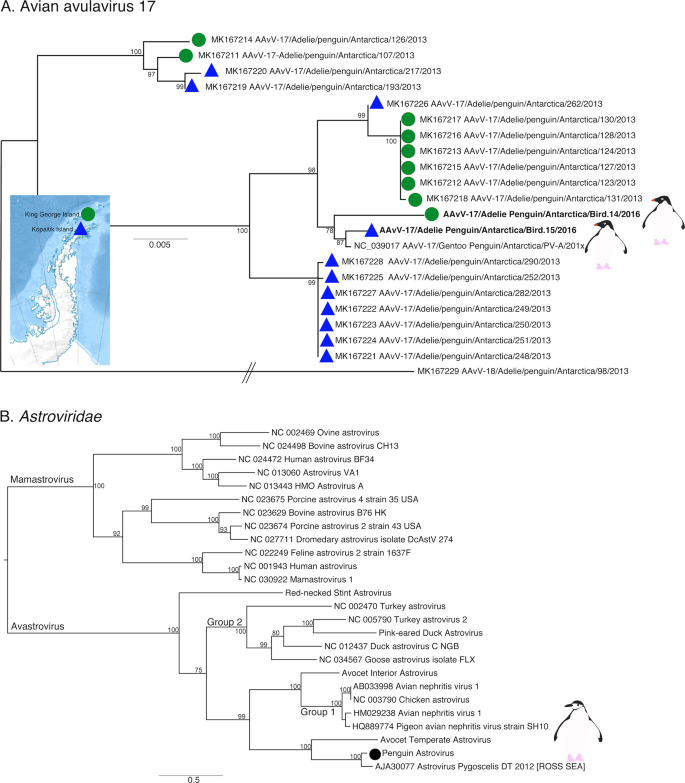
a Phylogeny of the F gene of Avian avulavirus 17. Detection location for viruses identified in this study and Wille et al. [21] are denoted by either a green filled circle (King George Island) or blue filled triangle (Kopaitik Island). Strain names for reference viruses are as presented with the same nomenclature as originally presented in Wille et al. [21]. Avian avulavirus 18 was used as outgroup to root the tree. The scale bar represents the number of nucleotide substitutions per site. b Phylogenetic tree of the ORF1ab, including the RdRp, of avastroviruses. The tree is mid-point rooted for clarity only. The scale bar represents the number of amino acid substitutions per site. Bootstrap values > 70% are shown for key nodes. Viruses identified in this study are denoted in bold.
We also identified a deltacoronavirus and an avastrovirus (Figs. 6b and S10–S12). The deltacoronavirus was similar to those reported in birds in the United Arab Emirates, Australia, Niger, and Finland, with ~95% identity. A lack of sampling makes it challenging to determine how deltacoronaviruses in Antarctica and other continents may be shared (Figs. S10, S11). The astrovirus detected was similar (88.3% identity) to a short fragment (1000 bp) previously reported in Adelie penguins on the Ross ice shelf of Antarctica [22] (Table S2), a pattern confirmed by phylogenetic analysis (Fig. 6b). Phylogenetic analysis also reveals that this virus falls in an outgroup to Group 2 viruses, including Avian Nephritis virus (Figs. 6b and S12). Although we were unable to determine the epidemiology of these viruses in Antarctica, repeated detection on opposite ends of the Antarctic continent makes it possible that this is a penguin specific virus.
Tick associated viruses
The most abundant virus identified within the I. uriae ticks sampled here was a variant of Taggert virus, a nairovirus (order Bunyavirales) previously identified in penguin associated ticks on Macquarie Island: the contigs identified in our data showed 81.6% nucleotide sequence similarity in the RdRp region to Taggert virus [25] (Fig. 7). This Taggert virus variant accounted for 2.0% of total reads (87% of viral reads) in the adult female library and was found in all tick libraries. Because the nucleotide sequences of Taggert virus differed between libraries it is unlikely that they represent cross-library contamination. In addition, we identified 75 reads of Taggert virus in the library containing samples from Chinstrap penguins on Kopaitik Island. Importantly, the tick and penguin libraries were not sequenced on the same lane, or even in the same time frame, thereby excluding contamination. Two other members of the order Bunyavirales were also discovered—Ronne virus and Barre virus—both members of the Phenuiviridae (Fig. S13) that exhibited 80% amino acid sequence similarity across the RdRp segment. Ronne virus was identified in three of the tick libraries (adult male, adult female, and nymph library1) but Barre virus was identified only in a single library (nymph library 2) (Fig. 4).
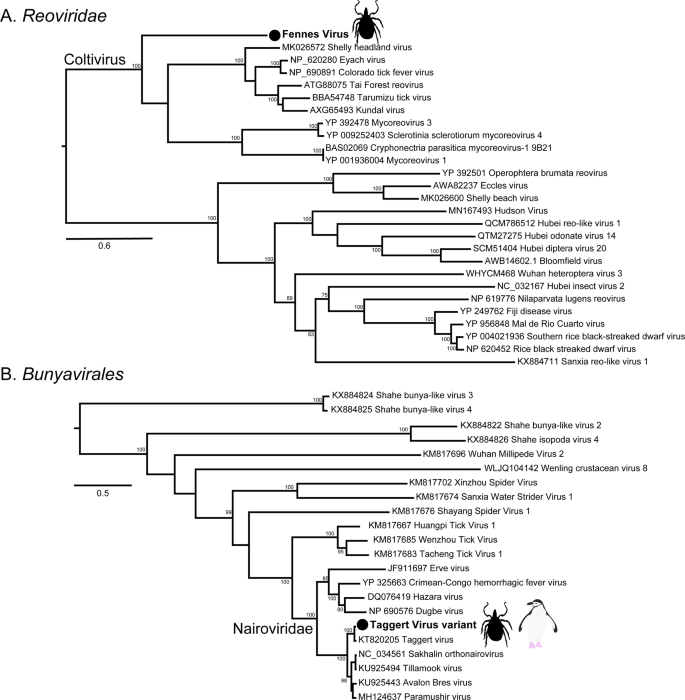
a The RdRp segment of select members of the Reoviridae, including the genus Coltivirus. b The RdRp of select members of the Bunyavirales including the family Nairoviridae. The novel tick viruses identified in this study are denoted with a filled circle and in bold. The tree has been mid-point rooted for clarity only. Bootstrap values >70% are shown for key nodes. The scale bar represents the number of amino acid substitutions per site.
The six other novel virus species identified in the tick libraries comprised five viral families: Iflaviridae-like, Alphatetraviridae, Reoviridae, Rhabdoviridae, and Levivirdae. A novel Ifla-like virus, Gerbovich virus, was identified within both nymph libraries. This virus clustered with a group of tick associated ifla-like viruses, including Ixodes holocyclus iflavirus and Ixodes scapularis iflavirus (Fig. S13). Two sister species of virus were identified within the Alphatetraviridae—Bulatov virus and Vovk virus—that showed 76.1% amino acid sequence similarity across the RdRp region. These two viruses are highly divergent from all other RdRp sequences currently available, exhibiting just 35.7% amino acid sequence similarity to the divergent tick-borne tetravirus-like virus (Fig. S13). A novel colti-like virus (Reoviridae), Fennes virus, was identified in the adult male, female and nymph libraries, although we were only able to assemble four segments. Notably, Fennes virus falls basal to the existing coltivirus group, exhibiting just 30% amino acid sequence similarity to Shelly headland virus, recently identified in I. holocyclus ticks from Australia (Fig. 7). The partial genome of a Rhabdovirus, Messner virus, was identified in the adult female library. However, this fragment was of low abundance, and only the RdRp segment (Fig. S13).
Finally, Mackintosh virus, identified in all four tick libraries, was not associated with any other tick viruses. Instead, this virus clustered with viruses from the Leviviridae indicating that it is likely a bacteriophage (Table S2).
Source: Ecology - nature.com


