Rohwer, F. & Thurber, R. V. Viruses manipulate the marine environment. Nature 459, 207–212 (2009).
Breitbart, M., Bonnain, C., Malki, K. & Sawaya, N. A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766 (2018).
Stewart, F. M. & Levin, B. R. The population biology of bacterial viruses: why be temperate. Theor. Popul. Biol. 26, 93–117 (1984).
Jiang, S. & Paul, J. Significance of lysogeny in the marine environment: studies with isolates and a model of lysogenic phage production. Microb. Ecol. 35, 235–243 (1998).
Howard-Varona, C., Hargreaves, K. R., Abedon, S. T. & Sullivan, M. B. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520 (2017).
Knowles, B. et al. Lytic to temperate switching of viral communities. Nature 531, 466–470 (2016).
Kang, H. S. et al. Prophage genomics reveals patterns in phage genome organization and replication. Preprint at bioRxiv https://doi.org/10.1101/114819 (2017).
Zhao, Y. et al. Abundant SAR11 viruses in the ocean. Nature 494, 357–360 (2013).
Våge, S., Storesund, J. E. & Thingstad, T. F. SAR11 viruses and defensive host strains. Nature 499, E3–E4 (2013).
Giovannoni, S., Temperton, B. & Zhao, Y. Giovannoni et al. reply. Nature 499, E4–E5 (2013).
Thingstad, T. F. & Bratbak, G. Microbial oceanography: viral strategies at sea. Nature 531, 454–455 (2016).
Wigington, C. H. et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 1, 15024 (2016).
Parikka, K. J., Le Romancer, M., Wauters, N. & Jacquet, S. Deciphering the virus‐to‐prokaryote ratio (VPR): insights into virus–host relationships in a variety of ecosystems. Biol. Rev. 92, 1081–1100 (2017).
Knowles, B. & Rohwer, F. Knowles & Rohwer reply. Nature 549, E3–E4 (2017).
Weitz, J. S., Beckett, S. J., Brum, J. R., Cael, B. & Dushoff, J. Lysis, lysogeny and virus–microbe ratios. Nature 549, E1–E3 (2017).
Morris, R. M. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002).
Giovannoni, S. J. SAR11 bacteria: the most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 9, 231–255 (2017).
Jiang, S. C. & Paul, J. H. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar. Ecol. Prog. Ser. 104, 163–172 (1994).
Jiang, S. C. & Paul, J. H. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142, 27–38 (1996).
Leitet, C., Riemann, L. & Hagström, Å. Plasmids and prophages in Baltic Sea bacterioplankton isolates. J. Mar. Biol. Assoc. UK 86, 567–575 (2006).
Paul, J. H. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2, 579–589 (2008).
Durham, B. P. et al. Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat. Microbiol. 4, 1706–1715 (2019).
Rappe, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002).
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L. & Field, K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63 (1990).
Giovannoni, S. J. et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438, 82–85 (2005).
Giovannoni, S. J. et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245 (2005).
Fogg, P. C., Colloms, S., Rosser, S., Stark, M. & Smith, M. C. New applications for phage integrases. J. Mol. Biol. 426, 2703–2716 (2014).
Ptashne, M. Principles of a switch. Nat. Chem. Biol. 7, 484–487 (2011).
Owen, S. V. et al. Characterization of the prophage repertoire of African Salmonella Typhimurium ST313 reveals high levels of spontaneous induction of novel phage BTP1. Front. Microbiol. 8, 235 (2017).
Hay, I. D. & Lithgow, T. Filamentous phages: masters of a microbial sharing economy. EMBO Rep. 20, e47427 (2019).
Weitz, J. S., Li, G., Gulbudak, H., Cortez, M. H. & Whitaker, R. J. Viral invasion fitness across a continuum from lysis to latency. Virus Evol. 5, vez006 (2019).
Toyofuku, M., Nomura, N. & Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24 (2019).
Biller, S. J. et al. Bacterial vesicles in marine ecosystems. Science 343, 183–186 (2014).
Biller, S. J. et al. Membrane vesicles in seawater: heterogeneous DNA content and implications for viral abundance estimates. ISME J. 11, 394–404 (2017).
Carini, P., Steindler, L., Beszteri, S. & Giovannoni, S. J. Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 7, 592–602 (2013).
Bondy-Denomy, J. et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 10, 2854–2866 (2016).
Ofir, G. & Sorek, R. Vesicles spread susceptibility to phages. Cell 168, 13–15 (2017).
Mizuno, C. M., Rodriguez-Valera, F., Kimes, N. E. & Ghai, R. Expanding the marine virosphere using metagenomics. PLoS Genet. 9, e1003987 (2013).
Beaulaurier, J. et al. Assembly-free single-molecule nanopore sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 30, 437–446 (2020).
Deschamps, P., Zivanovic, Y., Moreira, D., Rodriguez-Valera, F. & López-García, P. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic Thaumarchaeota and Euryarchaeota. Genome Biol. Evol. 6, 1549–1563 (2014).
Chen, L. X. et al. Wide distribution of phage that infect freshwater SAR11 bacteria. mSystems 4, e00410-19 (2019).
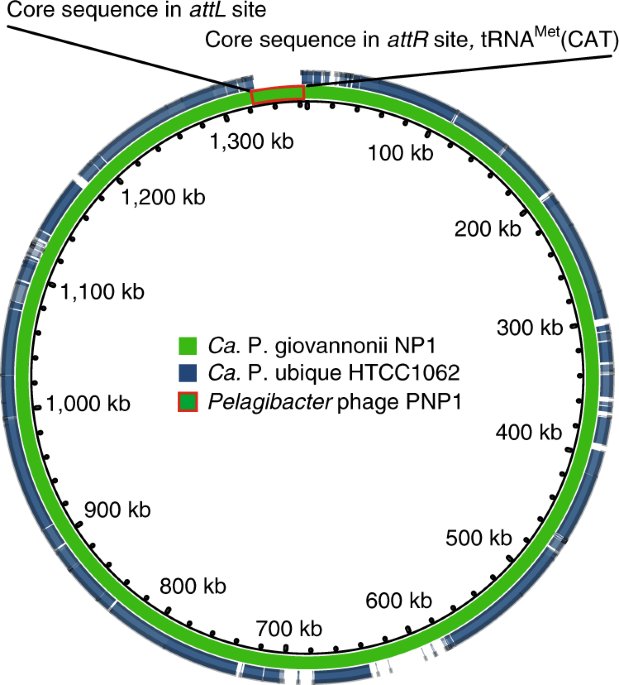
Zhao, Y. et al. Pelagiphages in the Podoviridae family integrate into host genomes. Environ. Microbiol. 21, 1989–2001 (2019).
Chin, C. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Alikhan, N., Petty, N. K., Zakour, N. L. B. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Noble, R. T. & Fuhrman, J. A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14, 113–118 (1998).
Source: Ecology - nature.com


