Erickson, J. D. Endangering the economics of extinction. Wildl. Soc. Bull. 28, 34–41 (2000).
Peres, C. A. Overharvesting. In Conservation biology for all (eds. Sodhi, N. S. & Ehrlich, P. R.) (Oxford University Press, 2010).
IUCN red list of threatened species: a global species assessment. (IUCN, 2004). 2004
Farrow, S. Extinction and market forces: two case studies. Ecol. Econ. 13, 115–123 (1995).
Worm, B. et al. Rebuilding global fisheries. Science 325, 578–585 (2009).
Hutchings, J. A. Collapse and recovery of marine fishes. Nature 406, 882–885 (2000).
Worm, B. et al. Impacts of biodiversity loss on ocean ecosystem services. Science 314, 787–790 (2006).
Ceballos, G., Ehrlich, P. R. & Dirzo, R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. USA 114, E6089–E6096 (2017)
Lande, R. Genetics and demography in biological conservation. Science 241, 1455–1460 (1988).
Shaffer, M. L. Minimum population sizes for species conservation. BioScience 31, 131–134 (1981).
Frankham, R. Genetics and extinction. Biol. Conserv. 126, 131–140 (2005).
Spielman, D., Brook, B. W. & Frankham, R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA 101, 15261–15264 (2004).
Harris, R. B., Wall, W. A. & Allendorf, F. W. Genetic consequences of hunting: what do we know and what should we do? Wildl. Soc. Bull. 30, 634–643 (2002).
Li, H. et al. Large numbers of vertebrates began rapid population decline in the late 19th century. Proc. Natl. Acad. Sci. USA 113, 14079–14084 (2016).
Nei, M., Maruyama, T. & Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975).
Tajima, F. The effect of change in population size on DNA polymorphism. Genetics 123, 597–601 (1989).
Excoffier, L., Dupanloup, I., Huerta-Sánchez, E., Sousa, V. C. & Foll, M. Robust demographic inference from genomic and SNP data. Plos Genet. 9, e1003905 (2013).
Hudson, R. R. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002).
Kingman, J. F. C. The coalescent. Stoch. Process. Their Appl. 13, 235–248 (1982).
Stoffel, M. A. et al. Demographic histories and genetic diversity across pinnipeds are shaped by human exploitation, ecology and life-history. Nat. Commun. 9, 4836 (2018).
Peery, M. Z. et al. Reliability of genetic bottleneck tests for detecting recent population declines. Mol. Ecol. 21, 3403–3418 (2012).
Beaumont, M. A., Zhang, W. & Balding, D. J. Approximate Bayesian Computation in population genetics. Genetics 162, 2025–2035 (2002).
Csilléry, K., Blum, M. G. B., Gaggiotti, O. E. & François, O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 25, 410–418 (2010).
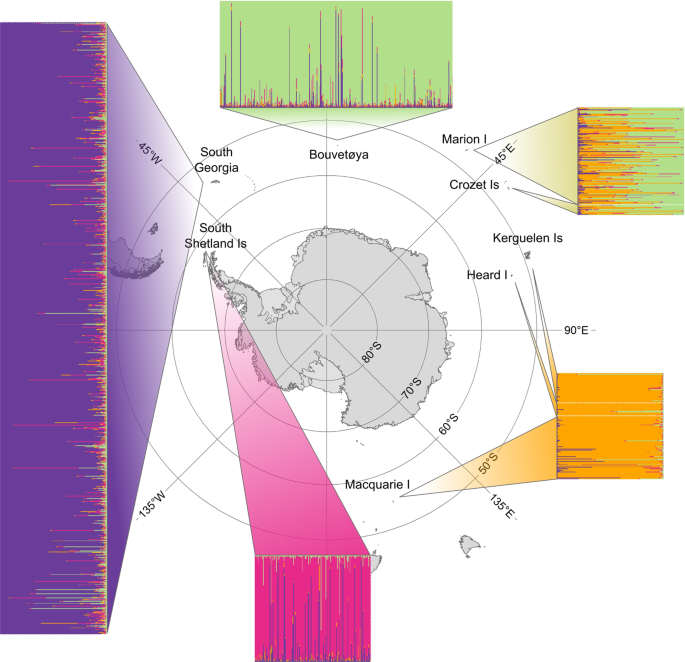
Hoffman, J. I., Boyd, I. L., Amos, W. & Ashley, M. Male reproductive strategy and the importance of maternal status in the Antarctic fur seal Arctocephalus gazella. Evolution 57, 1917–1930 (2003).
Hoffman, J. I., Trathan, P. N. & Amos, W. Genetic tagging reveals extreme site fidelity in territorial male Antarctic fur seals Arctocephalus gazella. Mol. Ecol. 15, 3841–3847 (2006).
Hoffman, J. I. & Forcada, J. Extreme natal philopatry in female Antarctic fur seals (Arctocephalus gazella). Mamm. Biol. – Z. Für Säugetierkd. 77, 71–73 (2012).
Forcada, J., Trathan, P. N. & Murphy, E. J. Life history buffering in Antarctic mammals and birds against changing patterns of climate and environmental variation. Glob. Change Biol. 14, 2473–2488 (2008).
Bonner, W. N. The fur seal of South Georgia. British Antarctic Survey-Scientific reports No. 56, 1–81 (1968).
Headland, R. K. A chronology of Antarctic exploration: a synopsis of events and activities from the earliest times until the international polar years, 2007-09. (Bernard Quaritch Ltd., 2009).
Mill, H. R. The siege of the South Pole: the story of Antarctic exploration. (Alston Rivers, Limited, 1905).
Fanning, E. Voyages & discoveries in the South Seas, 1792–1832. (Marine Research Society, 1833).
Weddell, J. A voyage towards the South Pole: performed in the years 1822-24. (London: Longman, Hurst, Rees, Orme, Brown, and Green, 1825).
Rand, R. W. Notes on the Marion Island fur seal. Proc. Zool. Soc. Lond. 126, 65–82 (1956).
Ingham, S. E. The status of seals (Pinnipedia) at Australian Antarctic stations. Mammalia 24, 422–430 (1960).
Shaughnessy, P. D. & Fletcher, L. Fur seals, Arctocephalus spp., at Macquarie Island. In Status, biology, and ecology of fur seals. Proceedings of an international symposium and workshop, Cambridge, England, 23–27 april 1984 177–188 (NOAA Technical Report NMFS 51, 1987).
Olstad, O. Trekk av Sydishavets dyreliv (Features of the Southern Ocean wildlife). Nor. Geogr. Tidsskr.-Nor. J. Geogr. 2, 511–534 (1928).
Payne, M. R. Growth of a fur seal population. Philos. Trans. R. Soc. Lond. B Biol. Sci. 279, 67–79 (1977).
Boyd, I. L. Pup production and distribution of breeding Antarctic fur seals (Arctocephalus gazella) at South Georgia. Antarct. Sci. 5, 17–24 (1993).
O’Gorman, F. A. Fur seals breeding in the Falkland Islands Dependencies. Nature 192, 914–916 (1961).
Condy, P. R. Distribution, abundance and annual cycle for fur seals (Arctocephalus spp.) on the Prince Edward Islands. South Afr. J. Wildl. Res. 8, 159–168 (1978).
Jouventin, P. & Weimerskirch, H. Long-term changes in seabird and seal populations in the Southern Ocean. In Antarctic ecosystems (eds. Kerry, K. R. & Hempel, G.) (Springer, 1990).
Jouventin, P. & Stonehouse, B. Biological survey of Ile de Croy, Iles Kerguelen, 1984. Polar Rec. 22, 688–691 (1985).
Budd, G. M. & Downes, M. C. Population increase and breeding in the Kerguelen fur seal, Arctocephalus tropicalis gazella, at Heard Island. Mammalia 33, 58–67 (1969).
Goldsworthy, S. D. et al. Fur seals at Macquarie Island: post-sealing colonisation, trends in abundance and hybridisation of three species. Polar Biol. 32, 1473–1486 (2009).
Kerley, G. I. H. Relative population sizes and trends, and hybridization of fur seals Arctocephalus tropicalis and A. gazella at the Prince Edward Islands, Southern Ocean. Afr. Zool. 18, 388–392 (1983).
Laws, R. M. Population increase of fur seals at South Georgia. Polar Rec. 16, 856–858 (1973).
Cleary, A. C. et al. Prey differences drive local genetic adaptation in Antarctic fur seals. Mar. Ecol. Prog. Ser. 628, 195–209 (2019).
Wynen, L. P. et al. Postsealing genetic variation and population structure of two species of fur seal (Arctocephalus gazella and A. tropicalis). Mol. Ecol. 9, 299–314 (2000).
Bonin, C. A., Goebel, M. E., Forcada, J., Burton, R. S. & Hoffman, J. I. Unexpected genetic differentiation between recently recolonized populations of a long-lived and highly vagile marine mammal. Ecol. Evol. 3, 3701–3712 (2013).
Humble, E. et al. RAD sequencing and a hybrid Antarctic fur seal genome assembly reveal rapidly decaying linkage disequilibrium, global population structure and evidence for inbreeding. G3 Genes Genomes Genet. 8, 2709–2722 (2018).
Hoffman, J. I. et al. A global cline in a colour polymorphism suggests a limited contribution of gene flow towards the recovery of a heavily exploited marine mammal. R. Soc. Open Sci. 5, 181227 (2018).
Gelman, A. et al. Bayesian data analysis. (Chapman and Hall/CRC, 2013).
Excoffier, L. & Foll, M. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27, 1332–1334 (2011).
Hoffman, J. I., Grant, S. M., Forcada, J. & Phillips, C. D. Bayesian inference of a historical bottleneck in a heavily exploited marine mammal. Mol. Ecol. 20, 3989–4008 (2011).
Shaughnessy, P. D. & Goldsworthy, S. D. Population size and breeding season of the Antarctic fur seal Arctocephalus gazella at Heard Island-1987/88. Mar. Mammal Sci. 6, 292–304 (1990).
Wege, M. et al. Trend changes in sympatric Subantarctic and Antarctic fur seal pup populations at Marion Island, Southern Ocean. Mar. Mammal Sci. 32, 960–982 (2016).
Hoban, S. M., Gaggiotti, O. E. & Bertorelle, G. The number of markers and samples needed for detecting bottlenecks under realistic scenarios, with and without recovery: a simulation-based study. Mol. Ecol. 22, 3444–3450 (2013).
Hahn, M. W. Molecular population genetics. (Sinauer Associates, 2018).
Frankham, R. Effective population size/adult population size ratios in wildlife: a review. Genet. Res. 66, 95–107 (1995).
Grafton, R. Q., Kompas, T. & Hilborn, R. W. Economics of overexploitation revisited. Science 318, 1601–1601 (2007).
Basberg, B. & Headland, R. K. The 19th century Antarctic sealing industry: sources, data and economic significance. (2008).
Stackpole, E. A. The Sea-Hunters: the New England whalemen during two centuries, 1635–1835. vol. 23 (J.B. Lippincott & Co., 1954).
Stackpole, E. A. The voyage of the Huron and the Huntress: the American sealers and the discovery of the continent of Antarctica. (Marine Historical Association, 1955).
O’Brien, S. J. A role for molecular genetics in biological conservation. Proc. Natl. Acad. Sci. USA 91, 5748–5755 (1994).
Hoelzel, A. R. Impact of population bottlenecks on genetic variation and the importance of life-history; a case study of the northern elephant seal. Biol. J. Linn. Soc. 68, 23–39 (1999).
Groombridge, J. J., Bruford, M. W., Jones, C. G. & Nichols, R. A. Evaluating the severity of the population bottleneck in the Mauritius kestrel Falco punctatus from ringing records using MCMC estimation. J. Anim. Ecol. 70, 401–409 (2001).
Hoelzel, A. R., Fleischer, R. C., Campagna, C., Le Boeuf, B. J. & Alvord, G. Impact of a population bottleneck on symmetry and genetic diversity in the northern elephant seal. J. Evol. Biol. 15, 567–575 (2002).
Amos, W. & Balmford, A. When does conservation genetics matter? Heredity 87, 257–265 (2001).
Amos, W. & Harwood, J. Factors affecting levels of genetic diversity in natural populations. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 353, 177–186 (1998).
Murray, G. G. R. et al. Natural selection shaped the rise and fall of passenger pigeon genomic diversity. Science 358, 951–954 (2017).
Dickerson, B. R., Ream, R. R., Vignieri, S. N. & Bentzen, P. Population structure as revealed by mtDNA and microsatellites in Northern fur seals, Callorhinus ursinus, throughout their range. Plos One 5, e10671 (2010).
Lancaster, M. L., Arnould, J. P. Y. & Kirkwood, R. Genetic status of an endemic marine mammal, the Australian fur seal, following historical harvesting. Anim. Conserv. 13, 247–255 (2010).
Goldsworthy, S., Francis, J., Boness, D. & Fleischer, R. Variation in the mitochondrial control region in the Juan Fernández fur seal (Arctocephalus philippii). J. Hered. 91, 371–377 (2000).
Majluf, P. & Goebel, M. E. The capture and handling of female South American fur seals and their pups. Mar. Mammal Sci. 8, 187–190 (2006).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning. vol. 2 (Cold spring harbor laboratory press New York, 1989).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Besnier, F. & Glover, K. A. ParallelStructure: a R package to distribute parallel runs of the population genetics program STRUCTURE on multi-core computers. Plos One 8, e70651 (2013).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Francis, R. M. pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32 (2017).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Goudet, J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (2005).
Archer, F. I., Adams, P. E. & Schneiders, B. B. stratag: An R package for manipulating, summarizing and analysing population genetic data. Mol. Ecol. Resour. 17, 5–11 (2016).
Keenan, K., McGinnity, P., Cross, T. F., Crozier, W. W. & Prodöhl, P. A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 4, 782–788 (2013).
Kamvar, Z. N., Tabima, J. F. & Grünwald, N. J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014).
Piry, S., Luikart, G. & Cornuet, J. M. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 90, 502–503 (1999).
Hofmeyr, G. J. G., Bester, M. N., Makhado, A. B. & Pistorius, P. A. Population changes in Subantarctic and Antarctic fur seals at Marion Island. South Afr. J. Wildl. Res. 36, 55–68 (2006).
Pacifici, M. et al. Generation length for mammals. Nat. Conserv. 5, 87–94 (2013).
Garza, J. C. & Williamson, E. G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 10, 305–318 (2001).
Csilléry, K., François, O. & Blum, M. G. B. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 3, 475–479 (2012).
Paijmans, A. J. et al. Data from: The genetic legacy of extreme exploitation in a polar vertebrate. Zenodo https://doi.org/10.5281/zenodo.3585717 (2019).
Source: Ecology - nature.com


