Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–58.
Hügler M, Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci. 2011;3:261–89.
Evans MC, Buchanan BB, Arnon DI. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci USA. 1966;55:928–34.
Fuchs G, Stupperich E, Eden G. Autotrophic CO2 fixation in Chlorobium limicola. Evidence for the operation of a reductive tricarboxylic acid cycle in growing cells. Arch Microbiol. 1980;128:64–71.
Schauder R, Widdel F, Fuchs G. Carbon assimilation pathways in sulfate-reducing bacteria II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch Microbiol. 1987;148:218–25.
Shiba H, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch Microbiol. 1985;141:198–203.
Ivanovsky RN, Sintsov NV, Kondratieva EN. ATP-linked citrate lyase activity in the green sulfur bacterium Chlorobium limicola forma thiosulfatophilum. Arch Microbiol. 1980;128:239–41.
Aoshima M, Ishii M, Igarashi Y. A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol Microbiol. 2004;52:751–61.
Aoshima M, Ishii M, Igarashi Y. A novel enzyme, citryl-CoA lyase, catalysing the second step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol Microbiol. 2004;52:763–70.
Kanao T, Fukui T, Atomi H, Imanaka T. ATP-citrate lyase from the green sulfur bacterium Chlorobium limicola is a heteromeric enzyme composed of two distinct gene products. Eur J Biochem. 2001;268:1670–8.
Beh M, Strauss G, Huber R, Stetter K-O, Fuchs G. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch Microbiol. 1993;160:306–11.
Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36.
Thauer RK, Möller-Zinkhan D, Spormann AM. Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu Rev Microbiol. 1989;43:43–67.
Ito S, Koyama N, Osanai T. Citrate synthase from Synechocystis is a distinct class of bacterial citrate synthase. Sci Rep. 2019;9:6038.
Mall A, Sobotta J, Huber C, Tschirner C, Kowarschik S, Bačnik K, et al. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science. 2018;359:563–7.
Nunoura T, Chikaraishi Y, Izaki R, Suwa T, Sato T, Harada T, et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science. 2018;359:559–63.
Ragsdale SW. Stealth reactions driving carbon fixation. Science. 2018;359:517–8.
Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, et al. Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv Micro Physiol. 2011;59:1–100.
Röling WFM. The family Geobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. p. 157–72.
Mahadevan R, Bond DR, Butler JE, Esteve-Nuñez A, Coppi MV, Palsson BO, et al. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol. 2006;72:1558–68.
Segura D, Mahadevan R, Juárez K, Lovley DR. Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PLoS Comput Biol. 2008;4:e36.
Lovley DR, Chapelle FH. Deep subsurface microbial processes. Rev Geophys. 1995;33:365–81.
Morita M, Malvankar NS, Franks AE, Summers ZM, Giloteaux L, Rotaru AE, et al. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. MBio. 2011;2:e00159–11.
Rotaru A-E, Shrestha PM, Liu F, Shrestha M, Shrestha D, Embree M, et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci. 2013;7:408–15.
Holmes DE, Shrestha PM, Walker DJF, Dang Y, Nevin KP, Woodard TL, et al. Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils. Appl Environ Microbiol. 2017;83:e00223–17.
Ueki T, Nevin KP, Woodard TL, Aklujkar MA, Holmes DE, Lovley DR. Construction of a Geobacter strain with exceptional growth on cathodes. Front Microbiol. 2018;9:1512.
Bond DR, Lovley DR. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl Environ Microbiol. 2003;69:1548–55.
Gregory KB, Bond DR, Lovley DR. Graphite electrodes as electron donors for anaerobic respiration. Environ Microbiol. 2004;6:596–604.
Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol. 2001;67:3180–7.
Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–9.
Caccavo F, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol. 1994;60:3752–9.
Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, et al. Biofuels. Altered sterol composition renders yeast thermotolerant. Science. 2014;346:75–78.
Tremblay P-L, Höglund D, Koza A, Bonde I, Zhang T. Adaptation of the autotrophic acetogen Sporomusa ovata to methanol accelerates the conversion of CO2 to organic products. Sci Rep. 2015;5:16168.
Atsumi S, Wu T-Y, Machado IMP, Huang W-C, Chen P-Y, Pellegrini M, et al. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol. 2010;6:449.
Conrad TM, Lewis NE, Palsson BØ. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol. 2011;7:509.
Zhang T, Shi X-C, Xia Y, Mai L, Tremblay P-L. Escherichia coli adaptation and response to exposure to heavy atmospheric pollution. Sci Rep. 2019;9:10879.
Dragosits M, Mattanovich D. Adaptive laboratory evolution – principles and applications for biotechnology. Micro Cell Fact. 2013;12:64.
Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–69.
Wiser MJ, Ribeck N, Lenski RE. Long-term dynamics of adaptation in asexual populations. Science. 2013;342:1364–7.
Tremblay P-L, Summers ZM, Glaven RH, Nevin KP, Zengler K, Barrett CL, et al. A c-type cytochrome and a transcriptional regulator responsible for enhanced extracellular electron transfer in Geobacter sulfurreducens revealed by adaptive evolution. Environ Microbiol. 2011;13:13–23.
Smith JA, Tremblay P-L, Shrestha PM, Snoeyenbos-West OL, Franks AE, Nevin KP, et al. Going wireless: Fe(III) oxide reduction without pili by Geobacter sulfurreducens strain JS-1. Appl Environ Microbiol. 2014;80:4331–40.
Summers ZM, Ueki T, Ismail W, Haveman SA, Lovley DR. Laboratory evolution of Geobacter sulfurreducens for enhanced growth on lactate via a single-base-pair substitution in a transcriptional regulator. ISME J. 2012;6:975–83.
Lovley DR, Phillips EJ. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol. 1986;51:683–9.
Picard Tools – By Broad Institute. http://broadinstitute.github.io/picard/. Accessed 29 Aug 2019.
Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4.
Bushnell B, Rood J, Singer E. BBMerge – accurate paired shotgun read merging via overlap. PLoS ONE. 2017;12:e0185056.
Holmes DE, Nevin KP, O’Neil RA, Ward JE, Adams LA, Woodard TL, et al. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol. 2005;71:6870–7.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8.
Nevin KP, Kim B-C, Glaven RH, Johnson JP, Woodard TL, Methé BA, et al. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS ONE. 2009;4:e5628.
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–6.
Lloyd JR, Leang C, Hodges Myerson AL, Coppi MV, Cuifo S, Methe B, et al. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem J. 2003;369:153–61.
Geelhoed JS, Henstra AM, Stams AJM. Carboxydotrophic growth of Geobacter sulfurreducens. Appl Microbiol Biotechnol. 2016;100:997–1007.
Speers AM, Reguera G. Electron donors supporting growth and electroactivity of Geobacter sulfurreducens anode biofilms. Appl Environ Microbiol. 2012;78:437–44.
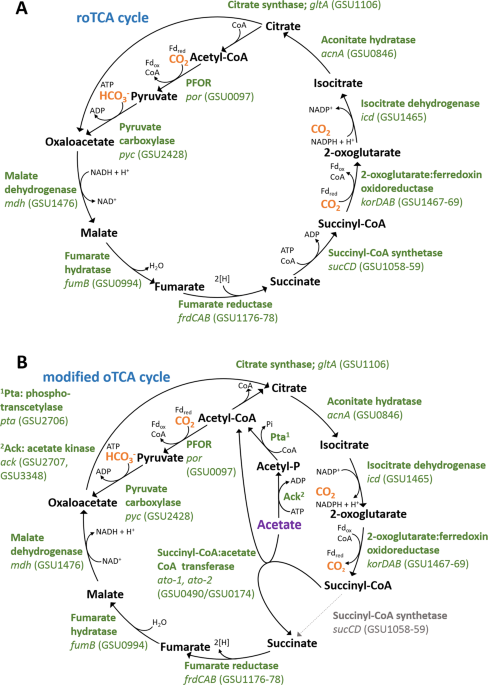
Galushko AS, Schink B. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch Microbiol. 2000;174:314–21.
Coppi MV, O’Neil RA, Lovley DR. Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J Bacteriol. 2004;186:3022–8.
Yang TH, Coppi MV, Lovley DR, Sun J. Metabolic response of Geobacter sulfurreducens towards electron donor/acceptor variation. Micro Cell Fact. 2010;9:90.
Coppi MV, O’neil RA, Leang C, Kaufmann F, Methé BA, Nevin KP, et al. Involvement of Geobacter sulfurreducens SfrAB in acetate metabolism rather than intracellular, respiration-linked Fe(III) citrate reduction. Microbiology. 2007;153:3572–85.
Härle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995;14:1430–8.
Coppi MV. The hydrogenases of Geobacter sulfurreducens: a comparative genomic perspective. Microbiology. 2005;151:1239–54.
You L, Page L, Feng X, Berla B, Pakrasi HB, Tang YJ. Metabolic pathway confirmation and discovery through (13)C-labeling of proteinogenic amino acids. J Vis Exp. 2012;59:3583.
Butler JE, Glaven RH, Esteve-Núñez A, Núñez C, Shelobolina ES, Bond DR, et al. Genetic characterization of a single bifunctional enzyme for fumarate reduction and succinate oxidation in Geobacter sulfurreducens and engineering of fumarate reduction in Geobacter metallireducens. J Bacteriol. 2006;188:450–5.
Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518.
LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30.
Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–61.
Rodríguez-Verdugo A, Carrillo-Cisneros D, González-González A, Gaut BS, Bennett AF. Different tradeoffs result from alternate genetic adaptations to a common environment. Proc Natl Acad Sci USA. 2014;111:12121–6.
Rodríguez-Verdugo A, Tenaillon O, Gaut BS. First-step mutations during adaptation restore the expression of hundreds of genes. Mol Biol Evol. 2016;33:25–39.
Rodríguez-Verdugo A, Gaut BS, Tenaillon O. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol Biol. 2013;13:50.
Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers are based in Hauppauge, New York, United States; 2011. p. 61–78.
Cha S. [13] Succinate thiokinase from pig heart: [EC 6.2.1.4. Succinate: CoA ligase (GDP)]. Method Enzymol. 1969;13:62–69.
Aklujkar M, Young ND, Holmes D, Chavan M, Risso C, Kiss HE, et al. The genome of Geobacter bemidjiensis, exemplar for the subsurface clade of Geobacter species that predominate in Fe(III)-reducing subsurface environments. BMC Genomics. 2010;11:490.
Feist AM, Nagarajan H, Rotaru A-E, Tremblay P-L, Zhang T, Nevin KP, et al. Constraint-based modeling of carbon fixation and the energetics of electron transfer in Geobacter metallireducens. PLoS Comput Biol. 2014;10:e1003575.
Lovley DR. Electrically conductive pili: Biological function and potential applications in electronics. Curr Opin Electrochem. 2017;4:190–8.
Source: Ecology - nature.com


