DNA extraction
Malayan tapir whole blood samples (n = 2) were collected from the National Zoo of Malaysia and Sungai Dusun WCC, PERHILITAN. Dried blood spots (DBS, n = 19), tissues (n = 22), and hair samples (n = 6) were obtained from the Wildlife Genomic Resource Bank (WGRB) of the National Wildlife Forensic Laboratory, PERHILITAN (permit ref. NRE 600-2/2/21 JILID 2(42)). In addition, we included 23 faecal samples collected from 11 tapirs on different dates in 2014 and 2015 at the National Zoo of Malaysia (n = 8) and Sungai Dusun (n = 15); see Supplementary Table S2 for the detailed list of samples. The samples were stored in -20 °C within two hours of collection. All the sampling procedures for the whole blood samples were approved by the Institutional Animal Care and Use Committee, Universiti Putra Malaysia (ethical approval ref.: UPM/IACUC/AUP-R033/2016). All methods were performed in accordance with the Universiti Putra Malaysia Code of Practice for the Care and Use of Animals for Scientific Purposes. Genomic DNA (gDNA) was extracted using the QIAamp® DNA Mini Kit (Qiagen, Germany) following the manufacturer’s spin protocol for all the samples except the faeces, from which the gDNA was extracted using the E.Z.N.A® Soil DNA Kit (Omega Bio-tek, USA). Of the 72 samples, 54 samples including 23 faecal samples with information on their sexes were used for marker testing, validation, and characterisation (see Supplementary Tables S2–S4 for details), while the other 18 samples of unknown sex were sex-typed (Table 1) to supplement sex ratio estimation.
Cross-amplification of Equus sp. Y-chromosome specific microsatellite markers
The six Y-specific microsatellite markers isolated from Equus sp., namely: Eca.YM2, Eca.YJ10, Eca.YH12, Eca.YE1, Eca.YP9 and Eca.YA1630, were tested for cross-amplification in four samples (2 males: 2 females). Singleplex PCR reactions were carried out in total volumes of 10 μL, each containing 1 × MyTaq Red Mix (Bioline, Germany), 10 ng of gDNA, and 0.5, 0.8 or 1.0 μM of each primer. The PCR was run using touchdown protocols, which included an initial denaturation at 95 °C for 1 min followed by 35 cycles of denaturation at 95 °C for 15 s, annealing at 59 → 49 °C (Eca.YM2), 61 → 51 °C (Eca.YJ10), 62 → 52 °C (Eca.YH12 and Eca.YE1), 58 → 47 °C (Eca.YP9), or 54 → 45 °C (Eca.YA16) for 15 s (−1 °C per cycle), and extension at 72 °C for 15 s, ending with a single cycle of final extension at 72 °C for 3 mins.
PCR amplification and characterisation of SRY and ZF genes
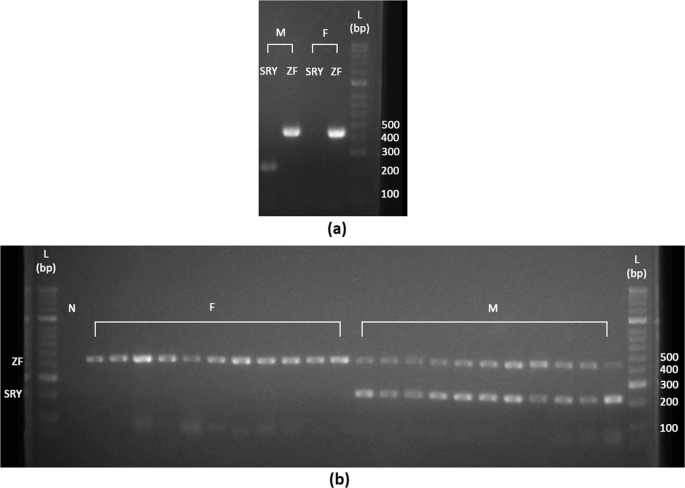
The primers for the amplification of the ZF gene (P1-5EZ: 5′-ATA ATC ACA TGG AGA GCC ACA AGC T-3′ and P2-3EZ: 5′-GCA CTT CTT TGG TAT CTG AGA AAG T-3′) and the SRY gene (Y53-3C: 5′-CCC ATG AAC GCA TTC ATT GTG TGG-3′ and Y53-3D: 5′-ATT TTA GCC TTC CGA CGA GGT CGA TA-3′) were taken from published literature32,33. Singleplex PCR reactions were performed in 10 μL mixture containing 1 × MyTaq Red Mix, 0.5 μM of P1-5EZ/P2-3EZ or Y53-3C/3D primer mix, and 2 ng of gDNA. The PCR was run using the touchdown profile: an initial denaturation at 95 °C for 3 mins followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 65 → 55 for 20 s (-1 °C per cycle) and extension at 72 °C for 20 s, and a final extension at 72 °C for 7 mins. For sex-typing, both SRY and ZF gene amplicons were expected in male samples, while only ZF gene amplicons were expected in female samples. The amplicons of SRY and ZFX from a male and a female respectively, were cloned into pGEM®-T Easy Vector (Promega, USA) following the manufacturer’s instructions and then sequenced to verify the amplified loci. Sequencing was run in both directions. The sequences were then searched against NCBI database using the BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The sex-typing primers were evaluated by PCR amplification in 22 samples (11 males: 11 females) consisting of tissue, whole blood, and DBS samples (Supplementary Table S4). A negative control without DNA template was included. The conditions of the PCR reactions remained the same for SRY/ZF method, but primers were multiplexed at 0.4 μM of P1-5EZ/P2-3EZ and 0.5 μM Y53-3C/3D. All the amplicons were electrophoresed through 2% agarose gel stained with RedSafeTM Nucleic Acid Staining Solution (20 ml gel: 1 μl staining solution), and the gel images were captured in an ENDUROTM GDS-1302 gel documentation system (Edison, NJ, USA).
Marker characterisation was carried out by modifying the primers using the method described by Vartia et al.44. The 5′-ends of P1-5EZ and Y53-3D were extended with the sequence of a third tail ‘Neomycin rev’ labelled with 6-FAM (5′6-FAM-AGG TGA GAT GAC AGG AGA TC-3′). The primer concentrations of the two markers were 0.4 μM for ZF and 0.5 μM for SRY. However, the ratio of the unmodified primer:tailed primer:Neomycin rev was adjusted to 4:1:4. PCRs were run using the same touchdown protocol for 18 samples (9 males:9 females), consisting of tissue, whole blood and DBS, that were chosen from different locations to look for possible length polymorphisms (Supplementary Table S5). Fragment analysis was performed on an ABI Genetic Analyzer ABI3730XL (Applied Biosystems, USA) using LIZ500 as the size standard. The peaks were examined and scored in Peak Scanner v2.0 (Applied Biosystems, Carlsbad, CA).
Since faeces is one of the most common non-invasive sources of DNA collected for wildlife species, the molecular sexing method was also tested in 23 samples of tapir faecal DNA following the same PCR profile using the unmodified primer pairs. Three PCR replicates were prepared per sample for sex-typing and evaluation.
Sex-typing samples of unknown sex
The 18 samples without information on their sexes were sex-typed either on 2% agarose gel, or by fragment analysis following the respective conditions described above, and in three PCR replicates per sample (Supplementary S3). Fragment analysis, a more sensitive assay, was only employed if the initial results on agarose gel were doubted due to the low quality and quantity of DNA samples that resulted in smearing, and low or no amplification. Fragment analysis results were used as a reference for sexing. In either method, band or peak patterns obtained in at least two out of three replicates and within the premise of successful amplification of ZF gene were accepted for sex-typing.
Preliminary assessment of sex ratio
In this study, sex ratio is defined as the proportion of male, expressed as male/(male + female), throughout the text and is denoted as a value between 0 (all females) and 1 (all males). For sex ratio estimation, tapir data was extracted from separate datasheets provided by National Wildlife Forensic Laboratory, PERHILITAN (WGRB Perissodactyla database), and Sungai Dusun WCC, PERHILITAN, and supplemented by information provided by National Zoo of Malaysia and Taiping Zoo. The WGRB database contained sex information of tapirs whose samples were collected and stored in the bank. The result of molecular sexing for samples with missing sex information described above were also used to supplement the dataset. Tapirs labelled as “wild” were tapirs encountered in the wild, including the rescued, translocated, trapped, and road-killed tapirs. Tapirs of unknown origin were those neither labelled as “wild” nor “captive”. Potential replicate entries from the same individuals were identified and merged using one or multiple identifiers in combination, including name, reference number, studbook number, microchip identification number, and descriptions in the “remarks” column. Captive-born tapirs and tapirs without both information on their sexes and biological samples for sexing were omitted.
An overall estimation of sex ratio was calculated in wild tapirs pooled with samples of unknown origin but presumably wild (in total N = 66, see Supplementary Table S6), and in wild tapirs alone (N = 47). Since sex, under normal conditions, has only two outcomes: male and female, the binomial test was used as the significant test for deviation from 1:1 ratio or a frequency of 0.5, carried out using the function binom.test in R.3.5.145 run in RStudio 1.1.44746.
Source: Ecology - nature.com


