Horton, T., Lowry, J. & De Broyer, C. World amphipoda database, http://www.marinespecies.org/amphipoda (2017).
Copilaș‐Ciocianu, D., Zimța, A. A. & Petrusek, A. Integrative taxonomy reveals a new Gammarus species (Crustacea, Amphipoda) surviving in a previously unknown southeast European glacial refugium. J. Zool. Syst. and Evol. Res. 57, 272–297 (2019).
Holsinger, J. R. Pattern and process in the biogeography of subterranean amphipods. Hydrobiologia 287, 131–145 (1994).
Jelassi, R., Khemaissia, H., Zimmer, M., Garbe-Schönberg, D. & Nasri-Ammar, K. Biodiversity of Talitridae family (Crustacea, Amphipoda) in some Tunisian coastal lagoons. Zool. Stud. 54, 17 (2015).
Romanova, E. V. et al. Evolution of mitochondrial genomes in Baikalian amphipods. BMC Genomics 17, 1016 (2016).
Tomikawa, K. & Nakano, T. Two new subterranean species of Pseudocrangonyx Akatsuka & Komai, 1922 (Amphipoda: Crangonyctoidea: Pseudocrangonyctidae), with an insight into groundwater faunal relationships in western Japan. J. Crustacean Biol. 38, 460–474 (2018).
Wildish, D. Reproductive consequences of the terrestrial habit in Orchestia (Crustacea: Amphipoda). Int. J. Invert. Reprod. 1, 9–20 (1979).
Griffiths, C., Stenton-Dozey, J. & Koop, K. In Sandy beaches as ecosystems 547–556 (Springer, 1983).
Duarte, C., Navarro, J., Acuña, K. & Gómez, I. Feeding preferences of the sandhopper Orchestoidea tuberculata: the importance of algal traits. Hydrobiologia 651, 291–303 (2010).
Rainbow, P., Malik, I. & O’brien, P. Physicochemical and physiological effects on the uptake of dissolved zinc and cadmium by the amphipod crustacean Orchestia gammarellus. Aquat. Toxicol. 25, 15–30 (1993).
Casini, S. & Depledge, M. Influence of copper, zinc, and iron on cadmium accumulation in the talitrid amphipod, Platorchestia platensis. Bull. Environ. Contam. and Toxicol. 59, 500–506 (1997).
Ungherese, G. et al. Relationship between heavy metals pollution and genetic diversity in Mediterranean populations of the sandhopper Talitrus saltator (Montagu)(Crustacea, Amphipoda). Environ. Pollut. 158, 1638–1643 (2010).
Bickham, J. W., Sandhu, S., Hebert, P. D., Chikhi, L. & Athwal, R. Effects of chemical contaminants on genetic diversity in natural populations: implications for biomonitoring and ecotoxicology. Mutat. Res. 463, 33–51 (2000).
De Wolf, H., Blust, R. & Backeljau, T. The population genetic structure of Littorina littorea (Mollusca: Gastropoda) along a pollution gradient in the Scheldt estuary (The Netherlands) using RAPD analysis. Sci. Total Environ. 325, 59–69 (2004).
Mohapatra, A., Rautray, T., Patra, A. K., Vijayan, V. & Mohanty, R. K. Elemental composition in mud crab Scylla serrata from Mahanadi estuary, India: in situ irradiation analysis by external PIXE. Food Chem. Toxicol. 47, 119–123 (2009).
Pavesi, L., Tiedemann, R., De Matthaeis, E. & Ketmaier, V. Genetic connectivity between land and sea: the case of the beachflea Orchestia montagui (Crustacea, Amphipoda, Talitridae) in the Mediterranean Sea. Front. Zool. 10, 21 (2013).
Ketmaier, V., Matthaeis, E. D., Fanini, L., Rossano, C. & Scapini, F. Variation of genetic and behavioural traits in the sandhopper Talitrus saltator (Crustacea Amphipoda) along a dynamic sand beach. Ethol. Ecol. Evol. 22, 17–35 (2010).
Fanini, L., Marchetti, G. M., Baczewska, A., Sztybor, K. & Scapini, F. Behavioural adaptation to different salinities in the sandhopper Talitrus saltator (Crustacea: Amphipoda): Mediterranean vs Baltic populations. Mar. Freshwat. Res. 63, 275–281 (2012).
Ugolini, A., Cincinelli, A., Martellini, T. & Doumett, S. Salt concentration and solar orientation in two supralittoral sandhoppers: Talitrus saltator (Montagu) and Talorchestia ugolinii Bellan Santini and Ruffo. J. Comp. Physiol. A 201, 455–460 (2015).
Nourisson, D. & Scapini, F. Seasonal variation in the orientation of Talitrus saltator on a Mediterranean sandy beach: an ecological interpretation. Ethol. Ecol. Evol. 27, 277–293 (2015).
Rivarola‐Duarte, L. et al. A first glimpse at the genome of the Baikalian amphipod Eulimnogammarus verrucosus. J. Exp. Zool. B: Mol. Dev. Evol. 322, 177–189 (2014).
Poynton, H. C. et al. The Toxicogenome of Hyalella azteca: A Model for Sediment Ecotoxicology and Evolutionary Toxicology. Environ. Sci. Technol. 52, 6009–6022 (2018).
Zeng, V. et al. De novo assembly and characterization of a maternal and developmental transcriptome for the emerging model crustacean Parhyale hawaiensis. BMC Genomics 12, 581 (2011).
Yo, Y. W. T. (Crustacea–Amphipoda) of the Korean coasts. Beaufortia 38, 153–178 (1988).
Kumar Patra, A. et al. The complete mitochondrial genome of the sand-hopper Trinorchestia longiramus (Amphipoda: Talitridae). Mitochon. DNA B 4, 2104–2105 (2019).
Woo, J. et al. Demographic history of Trinorchestia longiramus (Amphipoda, Talitridae) in South Korea inferred from mitochondrial DNA sequence variation. Crustaceana 89, 1559–1573 (2016).
Sasago, Y. Study for distribution and molecular phylogenetic analysis of the talitrid amphipods in Japan, M. Sc. Thesis, Mie University, Tsu, (2011).
Quevillon, E. et al. InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120 (2005).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: a laboratory manual. (Cold Spring Harbor Laboratory Press, 1989).
Woo, S. et al. Efficient isolation of intact RNA from the soft coral Scleronephthya gracillimum (Kükenthal) for gene expression analyses. Integr. Biosci. 9, 205–209 (2005).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
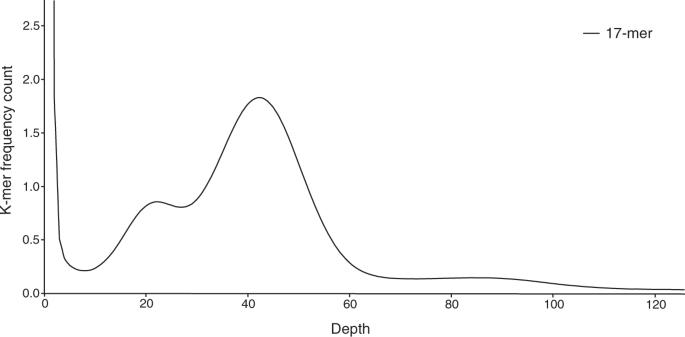
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Kajitani, R. et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395 (2014).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2010).
Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59 (2015).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Abrusán, G., Grundmann, N., DeMester, L. & Makalowski, W. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25, 1329–1330 (2009).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Bedell, J. A., Korf, I. & Gish, W. MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics 16, 1040–1041 (2000).
Hoff, K. J., Lange, S., Lomsadze, A., Borodovsky, M. & Stanke, M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769, https://doi.org/10.1093/bioinformatics/btv661 (2016).
Lomsadze, A., Burns, P. D. & Borodovsky, M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 42, e119–e119 (2014).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Minoche, A. E. et al. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 16, 184 (2015).
Au, K. F., Underwood, J. G., Lee, L. & Wong, W. H. Improving PacBio long read accuracy by short read alignment. Plos One 7, e46679, https://doi.org/10.1371/journal.pone.0046679 (2012).
Wu, T. D. & Watanabe, C. K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494 (2013).
Slater, G. S. C. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
She, R., Chu, J. S.-C., Wang, K., Pei, J. & Chen, N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19, 143–149 (2009).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Lima, T. et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 37, D471–D478 (2008).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2011).
Nikolskaya, A. N., Arighi, C. N., Huang, H., Barker, W. C. & Wu, C. H. PIRSF family classification system for protein functional and evolutionary analysis. Evol. Bioinform. 2, 117693430600200033 (2006).
Attwood, T. K. et al. PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res. 28, 225–227 (2000).
Bru, C. et al. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 33, D212–D215 (2005).
Sigrist, C. J. et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166 (2009).
Madera, M., Vogel, C., Kummerfeld, S. K., Chothia, C. & Gough, J. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32, D235–D239 (2004).
Haft, D. H. et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41, D387–D395 (2012).
Patra, A. K. et al. First draft genome for the sand-hopper Trinorchestia longiramus. figshare, https://doi.org/10.6084/m9.figshare.8217854.v5 (2020).
NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRP199018 (2019).
Patra, A. K. et al. Trinorchestia longiramus isolate TLONG-mixed, whole genome shotgun sequencing project. GenBank, https://identifiers.org/ncbi/insdc:VCRD00000000 (2020).
NCBI Assembly, https://identifiers.org/ncbi/insdc.gca:GCA_006783055.1 (2019).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Li, L., Stoeckert, C. J. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973, https://doi.org/10.1093/bioinformatics/btp348 (2009).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Hedges, S. B., Dudley, J. & Kumar, S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (2006).
Han, M. V., Thomas, G. W., Lugo-Martinez, J. & Hahn, M. W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987–1997, https://doi.org/10.1093/molbev/mst100 (2013).
Francia, M. E. et al. A Toxoplasma gondii protein with homology to intracellular type Na+/H+ exchangers is important for osmoregulation and invasion. Exp. Cell Res. 317, 1382–1396 (2011).
Dermauw, W. & Van Leeuwen, T. The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 45, 89–110 (2014).
Radulović, Ž., Porter, L. M., Kim, T. K. & Mulenga, A. Comparative bioinformatics, temporal and spatial expression analyses of Ixodes scapularis organic anion transporting polypeptides. Ticks Tick Borne Dis. 5, 287–298 (2014).
Source: Ecology - nature.com


