Tap cleaning
To assess tap cleaning agent and method efficacy: (1) a protocol was developed; (2) a system was built to group biofilms in taps and one trial run was completed; (3) four data collection runs, one for each of the four cleaning agents, were completed; and (4) data were analyzed.
Through partner discussions, a literature review, and author experience, four locally-available cleaning agents (bleach (sodium hypochlorite), boiled water, soapy water, and vinegar), and seven cleaning methods (flowing agent through tap for 60 s, soaking assembled for 60 s, soaking assembled for 5 min, soaking disassembled for 60 s, scrubbing with a bottle brush five times assembled, scrubbing with a brush five times disassembled, and scrubbing with a bottle brush five times and soaking disassembled for 5 min) were selected. A control of no cleaning was included. Test taps were manufactured by Tomlinson Industries (HFSLT Faucet 3/4″–16 UNF 1″ Long, Cleveland, Ohio, US) and consist of a PPE body and jam nut with two washers.
A system was built to continuously flow 18 L of E. coli-spiked Luria-Bertani (LB) broth through taps to grow biofilms (Fig. 6). The system consisted of three plastic 10 L buckets, with eight taps attached to each bucket (24 taps total). Taps were latched open and drained by gravity into an enclosed funnel and 20 L collection bucket. The three 20 L buckets drained via 3/4” braided hose into a centrifugal pump (Grundfos CM 3–4, Bjerringbro, Denmark), and liquid from all buckets mixed together. A pressure-reducing valve on the pump was used to hold pressure in the 1” discharge hose at 20 psi. From the discharge hose, the fluid was pumped to the top of the system and distributed via a manifold into each of the three 10 L buckets with taps. Manual ball valves on the manifold were tuned to ensure that flow rate into each 10 L bucket maintained a 1″ fluid head above the taps. This head produced a laminar flow regime (Reynolds number = 1899) and a draining flow rate of 1.5 L/min through each tap. The system was run once for optimization, after which cooling controls were added, including a: chiller pumping a mixture of 80% ethylene glycol and 20% water at 0 °C through copper tubing wrapped around the left bucket in the system; a cooler filled with ice packs on the return line; and a fan to cool the pump.
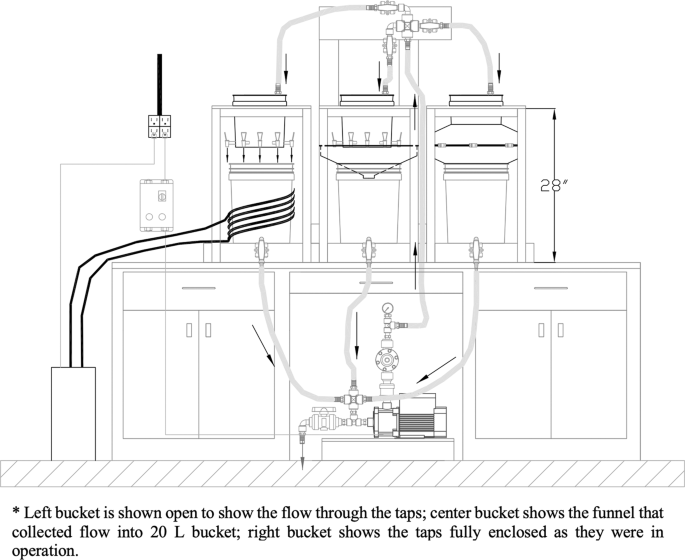
Schematic of the pumping system constucted in the laboratory for growing biofilm in taps*.
Four experimental runs were completed, each testing the efficacy of one of the cleaning agents using each of the cleaning methods. A run consisted of: sterilizing the pumping system; daily culture preparation; operation and monitoring of the system; cleaning agent preparation; cleaning and destruction of taps; microbiological sampling; and imaging.
Before starting an experimental run, the system was cleaned by removing buckets, taps, and funnels, and washing pieces with 0.5% sodium hypochlorite. After reassembly, the system was sterilized by pumping 5% sodium hypochlorite through all lines for 1 h, followed by one rinse cycle with 18 L sterile LB Broth, and rinse cycles with Type 1 laboratory-grade water (Milli-Q® Reference, MilliporeSigma, Burlington, MA, USA) filtered through a 0.22 µm filter (Millipak®, MilliporeSigma, Burlington, MA, USA), hereafter termed “Milli-Q”, to remove any residual chlorine. Confirmation that chlorine had been fully rinsed from the system was completed using a Lamotte 1200 colorimeter and DPD-1 instrument grade tablets (Lamotte, Chestertown, MD, USA) to ensure a non-detect chlorine residual before proceeding.
E. coli (ATCC 11229) stock was streaked onto LB agar plates, incubated at 35 °C, and stored at 4 °C. The night before each system refill, a streak plate colony was used to inoculate 20 mL of LB broth, and incubated at 35 °C on a rotating plate for 12–18 h with shaking. The culture was then diluted (1:20) in sterile LB broth and incubated at 35 °C on a rotating plate for 3 h, or until a concentration of ~1010 cells/mL was reach, as estimated using a spectrophotometer (OD = 600 nm). The volume of culture used to spike 102–103 CFU/mL E. coli into the sterile LB broth added to the pumping system was estimated from the spectrophotometer reading.
The system was operated by adding 6 L of E. coli-spiked LB broth to each of the 3 buckets (20 L each), pumping the broth through the system for 24 h, anddraining the system of the broth. This process was repeated for 5 days to grow biofilms on surfaces in contact with spiked broth. To monitor E. coli concentrations in the LB broth, an LB broth sample was aseptically collected at two time points: 15 min after adding fresh E. coli-spiked LB broth to the system; and, after 22.5 h of operation. Appropriate dilutions of these samples were prepared in phosphate buffered saline (PBS) (pH = 7.4), filtered through a 0.45 µm membrane, plated on mColiBlue24® (Hach, Loveland, CO) media, inverted and incubated at 35 °C for 24 h following standard methods. E. coli colonies were then enumerated and recorded. Please note we refer to this “E. coli-spiked LB broth” as “culture” herein. Culture temperature and volume were monitored twice daily during system operation, and 4 liters of Milli-Q water were added to the system every evening.
Preparation of cleaning agents is as follows. Bleach (0.5% sodium hypochlorite (NaOCl)) was prepared by diluting 6% laboratory-grade commercial bleach (Pure Bright, KIK International, Houston, TX, USA); concentration was confirmed using iodimetric titration (Hach Method 8209, Loveland, CO, USA). A 0.5% concentration was selected because: a 10:1 dilution of 5% commercial bleach is simple for a user to prepare; and 0.5% is recommended during Ebola outbreaks for disinfection of non-living surfaces, and has shown to efficaciously remove E. coli and viral surrogates from surfaces52. To prepare boiled water, Milli-Q water was dispensed into a 5 L metal cooking pot, placed on a hot plate, and brought to a roiling boiling. Before cleaning, water was poured into a sterilized 5 L graduated cylinder, from which water was dispensed as needed during cleaning. Please note water was not continuously boiling during cleaning but was checked to ensure it was ~80 °C. To prepare soapy water, commercially-available unbranded local bar soap was purchased in Dhaka, Bangladesh and shipped to Tufts University. Soap was grated with a kitchen cheese grater; 400 g of grated soap was then stirred into 20 L of Milli-Q water until mostly dissolved. This concentration was selected based on previous results finding this soapy water concentration was more effective at removing thermotolerant coliforms and C. perfringens from hands than water alone53. Lastly, commercially-available household vinegar (5% acetic acid) was purchased from a Boston-area grocery store and used without modification (Heinz, Pittsburgh, PA, USA).
At the conclusion of each experimental run, the system was drained and the 24 taps were unmounted. Three taps were cleaned using each of the seven cleaning methods; three control taps were not cleaned. Of the triplicate tap samples, two were swabbed for E. coli and one was destructed for imaging. To clean taps by flowing agents through them, taps were individually mounted on sterilized 10 L buckets, 4 L of agent was added to buckets and allowed to completely drain thought open spigots (approximately 60 s), then taps were aseptically unmounted. To soak taps, 200 mL of cleaning agent were added to sterile 300 mL Whirl-Pak® bags (Nasco, Fort Atkinson, WI, USA), a tap was added to the bag, bags were closed so taps were fully submerged, and after soaking, bags were drained and taps aseptically removed. To scrub taps, the sterile bottle brush was: (1) dipped into cleaning agent and the horizontal part was scrubbed in-and-out five times; and (2) dipped into cleaning agent and moved circularly around the inside surface of the horizontal part five times (Fig. 7). This scrubbing method was repeated for the vertical portion. To disassemble taps, the nut, two washers, and flow lever were unscrewed from the main body of the tap.
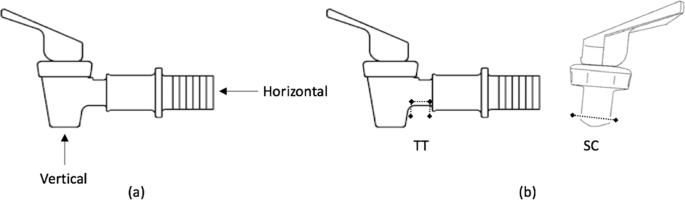
Tap scrubbing, swabbing, and coupon cutting locations, including a horizontal and vertical scrubbing and swabbing locations and b locations of the Tap Tube (TT) and the Seat Cup (SC) coupons removed for imaging54.
Swabbing was conducted using a Sanicult Hygiene Monitoring swab (Starplex Scientific, Etobicoke, Ontario, Canada). Each tap was swabbed separately in the horizontal and vertical directions by moving the swab ten times around the inner surface of the corresponding tap area (Fig. 7). The surface areas swabbed were ~31.45 cm2 and 4.59 cm2 for the horizontal and vertical surfaces, respectively. After swabbing, the swab was returned to the Sanicult broth, vortexed, and stored on ice. Appropriate dilutions of swab samples were prepared via membrane filtration, as described above.
One representative surface area (‘coupon’) was cut from each of two locations on each tap, the seat cup on the flow lever (SC) and the tap tube at the base of the tap’s horizontal section near the internal corner (TT) (Fig. 7). Each coupon was carefully cut from the tap using a sterile utility knife, rinsed with PBS, and allowed to air dry in a sterile field. Coupons were then mounted onto a glass microscope slide with nail polish and moved to a biosafety cabinet in a darkroom. In the dark, 50 µL of a 600 µM solution of 4’,6 diamidino-2-phenylindole dihydrochloride (DAPI) stain (MilliporeSigma, Burlington, MA, USA) were pipetted onto each coupon and incubated at room temperature for 30 min. Lastly, a drop of fluorescent mounting media (MilliporeSigma, Burlington, MA, USA) was applied as an anti-fading agent to each coupon, and a glass coverslip placed on top. Slides were wrapped in aluminum foil and stored at 4 °C until imaging.
Coupons were imaged by epifluorescence microscopy, using a LEICA SPE confocal microscope (Wetzlar, Germany) under ×63 objective in immersion oil. Images were acquired using a 405 nm visible laser diode. After locating surface E. coli, gain and offset were adjusted for each image to bring cells into focus. Three randomly selected fields of view were acquired for each coupon by scanning at 400 Hz from the top of the biofilm to the coupon’s surface. Image slices were recorded at predefined z-step sizes (ranging 0.3–2.0 µm). A resulting image stack (116.52 µm2 × depth of the sample) for each field of view was exported for analysis.
All collected microbiological data were entered into Microsoft Excel (Redmond, WA, USA). The geometric mean of plates in the countable range (10–200 colonies) was calculated for each swab after accounting for dilution, divided by the swabbed surface area, and reported as the number of colony forming units (CFU) per swabbed surface area (cm2). Results were averaged for each location for replicate taps.
Images were imported and visualized for qualitative analysis in the open-source software FIJI/ImageJ 2.0.0-rc-69/1.52n. They were converted to 8-bit black and white images, threheld using the Otsu algorithm, downsampled using a 3D Gaussian Blur algorithm (alpha = 0.5 all directions), then dilated once, eroded once, and inverted to remove background noise. Each image stack was then processed using the Particle Analyzer algorithm, E. coli counts and biofilm thickness recorded, and a density (CFU/µm3) calculated for each image stack. In denser biofilms E. coli cells clumped together to form colonies, which the algorithm counted as one, thus the calculated density value was sometimes artificially low. Thus, processed images were also qualitatively evaluated. A rating system of was developed to qualitatively categorize image stacks: no growth; a few disconnected E. coli cells and no clumping; a small amount of clumping and visually moderate biofilm structure; and large clumping and visually dense structure. All images were qualitatively classified by two trained individuals separately.
Surface container cleaning
To assess surface container cleaning efficacy: (1) a protocol was developed and coupons cut from containers; (2) one 21-day data collection run was completed, followed by a cleaning, E. coli testing, and image preparation; and (3) data were analyzed.
Polypropylene water storage containers were provided to Tufts University by partners. Using a scroll saw, 45 4 cm2 coupons were cut from the flat sides and bases of the containers. Using a Dektak XT-S Profilometer with Vision64 software (Bruker, Billerica, MA, USA), a surface profile was collected using a 12.5 µm tip stylus with 29.4 µN contact force at 166.7 µm/s scan speed by scanning each coupon for 5000 μm in three directions (x, y, and xy). Average roughness (Pa) and root-mean squared (Pka) values were calculated after leveling the profile using two points, and entered into Microsoft Excel. Outlier coupons were identified using the IQR and a trendline and removed from the sample set. The final sample size was 30 coupons of comparable surface roughness values (0.462–1.098 µm).
Coupons were sterilized with 0.5% bleach solution and 70% ethanol, and aseptically submerged in individual 50 mL Falcon tubes containing 25 mL of LB broth spiked with E. coli at a target concentration of 103 CFU/mL. E. coli culture was prepared as described in the tap study. Falcon tubes were incubated for 21 days at 35 °C on a ThermoFisher MaxQ 2000 orbital shaker (Fisher Scientific, Hampton, NH, USA) at 70 rpm. Coupons were aseptically transferred to new 50 mL Falcon tubes with 25 mL of fresh culture every 48 h, a total of 11 times throughout the 21-day experiment. Each spiking day, E. coli concentrations in culture were confirmed as in the tap study.
At the end of the 21-day growth period, the exterior surface of each coupon was sterilized with ethanol, rinsed by gently pipetting 2 mL of PBS across the interior surface to remove planktonic cells settled on the surface, and air dried in a sterile field. This ensured live E. coli in biofilm remained only on the interior PPE surface. Coupons were cleaned by applying three cleaning agents (0.53% bleach solution, 6% acetic acid vinegar, and boiled tap water) using three cleaning methods (soaking for five minutes, wiping with a sterile Whirl-Pak® hydrated PolySponge™ representative of a cleaning sponge (Nasco, Fort Atkinson, WI, USA), and wiping with a cloth (100″cm2, 60% cotton, 40% polyester). These methods were selected based on experience and results from the tap cleaning study. Before cleaning, cloth pieces were sterilized by soaking in bleach for 15 min, soaking in ethanol for 15 min, and drying in a biosafety cabinet for 18 h. A new, sterile sponge or cloth was used to clean each coupon. To apply the soaking cleaning method, a coupon was placed in a sterile 50 mL Falcon tube with 25 mL of cleaning agent, allowed to soak for 5 min, and removed from the Falcon tube. To apply the wiping cleaning methods, a coupon was placed interior facing upwards on a sterile surface, the sponge or cloth was soaked in the cleaning agent, and the coupon was wiped five times back and forth in each of four directions (xx, yy, xy, and yx). Please note, the same person wiped all coupons to maintain wiping consistency. Three replicate coupons were cleaned per agent/method combination, and three control coupons were left uncleaned (30 coupons total; three agents by three methods in triplicate, plus three controls).
After cleaning, two coupons from each triplicate were aseptically transferred into Falcon tubes containing 20 mL of PBS and if necessary a dissolved neutralizing agent (sodium thiosulfate for bleach, sodium bicarbonate for vinegar). Falcon tubes were vortexed on a Fisherbrand™ Analog Vortex Mixer for 30 s at 1200 rpm (Fisher Scientific, Hampton, NH, USA), then sonicated in a Fisherbrand™ FS20 Sonicator for 5 min at 40 Khz (Fisher Scientific, Hampton, NH, USA) in water at 20 °C. Falcon tubes samples were prepared as described in the above tap study to enumerate E. coli removed from surfaces via sonication. Additionally, one coupon from each triplicate was stained and imaged, as described above, with the exception the z-step size was maintained at 0.5 µm. Data were entered into Microsoft Excel and analyzed in Excel and R (RStudio, the R Foundation for Statistical Computing, Vienna, Austria).
Source: Resources - nature.com


