Study species
P. elegans is widely distributed throughout south-eastern and eastern Australia40. It is classified by the IUCN as of least concern due to its large range and high numbers, but is thought to be declining due to ongoing habitat destruction53. P. elegans use nest hollows for reproduction and show high nest site fidelity, but low social pair fidelity54. Female P. elegans can start breeding in their first year, whereas males typically only start breeding when they are at least two years old55. The species has been shown to be an excellent model system for BFDV research33,35,56.
Sample collection
We conducted this study under Deakin University animal ethics approval (B31-2015 and B37-2016) and Australian Bird and Bat Banding authority 2319, and it complied with the laws of Victoria (research permit 10007969). We collected samples from P. e. elegans in southern Victoria, Australia, from October 2016 until September 2018. We captured breeding birds during two breeding seasons using nest box traps57, from October 2016 to January 2017, and from October 2017 to January 2018. We also caught P. elegans in baited walk-in cage traps year-round (see Supplementary Table S1 and Supplementary Fig. S1 for information on numbers trapped, by age, sex, season and trap type). We used three field sites: Bellbrae (S38°19′ E144°11′, 110 nest boxes and two walk-in traps), Meredith/She Oaks (S37°51′ E144°06′, 50 nest boxes and two walk-in traps), and Steiglitz (S37°52′ E144°18′, 80 nest boxes and one walk-in trap).
We collected blood samples to study patterns of active infection (prevalence and intensity of infection)35, as well as cloacal swabs to estimate viral shedding32, and recorded body mass and tarsus length to estimate host condition51,58,59. We took ~100 µl of blood from the brachial vein and stored it in ethanol at room temperature. We took cloacal swabs and stored them at 3 °C in the field, then froze them at −80 °C upon return to the laboratory on the same day. To avoid virus transmission between sampled birds, the cotton bags we used to hold birds in were autoclaved after each use, blood sampling equipment was single-use and banding and measuring tools were sprayed with F10 SC Veterinary Disinfectant (Health and Hygiene Pty Ltd, South Africa) after each use.
We assigned birds to one of three age classes based on distinct plumage colouration: subadult (<1 year), young adult (1 – 3 years) and adult (≥3 years)56. Nestlings were not included in the study reported here. For analysis, subadults and young adults were combined into the age class ‘young bird’ (<3 years) to ensure an adequate sample size for comparison of breeding and non-breeding birds. To analyse changes in prevalence at a finer scale, we calculated age in months for these ‘young birds’, by calculating the estimated mean fledging time for the 2017 breeding season (n = 29 nests, mean 14 Dec, range 12 Nov – 17 Jan). We then correlated capture date with plumage colouration and wing stripe40, and assigned age in months accordingly (Supplementary Table S2). For example, we estimated that green subadults with full wing stripe on most or all primaries caught in January were one month old.
DNA extraction and PCR
To extract BFDV and host DNA from blood and swab samples, we used an ammonium acetate DNA extraction method that is commonly used in BFDV studies and gives high DNA yields12,33,60. The extracted DNA was stored in low Tris-EDTA buffer (10 mM Tris.HCL, 0.1 mM EDTA; pH 7.5 – 8.0) at -20 °C34. We determined DNA quality and quantity using a DU 640B spectrophotometer (Beckman Coulter, CA, U.S.A.) with a 1: 200 dilution. We sexed birds using a modified PCR protocol by Griffiths, et al.61. For BFDV detection, we diluted DNA to the same concentration (200 ng/µl), and then used a probe-based quantitative real-time PCR (qPCR) method34. We ran the assay using a PikoReal Real-Time PCR System (Thermo Fisher Scientific Inc., MA, U.S.A.). We added positive and no-template controls to each qPCR plate, and all samples were run in duplicate. Duplicate samples with Cq values (cycle at which probe fluorescence crosses the arbitrarily set detection baseline) differing by more than one cycle were run again. Although the qPCR method amplifies a shorter fragment of viral DNA than conventional PCR, it has been shown to deliver comparable results, while being more sensitive than the conventional assay34. qPCR is a widely used method for BFDV surveillance50. As most positives detected by qPCR represent active infection with viable virus as confirmed by sequencing33, but some can be non-active remnant viral DNA, we define prevalence as the percentage of individuals positive for BFDV viral DNA35. BFDV-positive (BFDV+) birds are termed either BFDV+blood or BFDV+cloacal, depending on which sample type was positive for BFDV presence. Samples that were BFDV-negative are termed BFDV−blood or BFDV−cloacal.
For comparative analysis of viral load across individuals, we re-ran BFDV+blood samples we used for comparative viral load analysis on the same qPCR plate, again all at a concentration of 200 ng/µl, to avoid possible slight variation of results across plates. We then used a comparative method to calculate viral load using the Cq values of each BFDV-positive sample33,62: Viral load = 2(−ΔCq). The resulting data were then log10-transformed to achieve normality34. Diluting cloacal swab DNA to the same concentration was not possible due to much lower DNA yield. We therefore only report prevalence for cloacal swabs, and not viral load. We re-tested a subset of walk-in-trapped P. elegans blood samples (subset with the highest BFDV prevalence) which were BFDV−blood, to estimate the likelihood of false negatives. None of the samples which were initially BFDV−blood came up BFDV+blood in the repeat run.
Statistical analyses
We carried out statistical analyses using SPSS 25.01 (IBM, Armonk NY, U.S.A.). To ensure independence of cases, we used only data from the first capture if individuals were recaptured. We pooled the data from our two years of study and from the three field sites, because we found overlapping confidence intervals between years and sites during initial data analysis (Supplementary Fig. S6). Additionally, Eastwood, et al.33 found that in P. elegans, geographic location, host density and parrot community diversity and composition do not explain differences in BFDV prevalence or load. A very low BFDV prevalence in some sex/age groups did not permit us to include year and trapping site as random intercepts, as well as age and sex in the same model, as the models did not converge. When we ran site and year separately as main effects in the full models, they were not significant, while we still found effects of age, date and sex (Supplementary Tables S7 – S10). We combined two of our three field sites, namely Meredith/She Oaks and Steiglitz, for statistical analysis. The two sites are located within 10 km of each other. Data from previous studies show that most recaptured P. elegans were trapped or resighted within approximately 10 km of their banding site54,63. We therefore defined Meredith/She Oaks and Steiglitz as one P. elegans population. We are aware that due to different and sometimes low sample sizes of infected birds trapped in some years and sites, and the resulting grouping of years and sites for most of our analyses, some of the observed seasonal patterns may in fact be stochastic effects. We address this in the discussion.
Initial calculations of prevalence included birds trapped during the breeding season (September – January, caught as parental birds in nest box, hereafter referred to as ‘breeding’) and birds trapped outside the breeding season (February – August, caught in walk-in traps, hereafter referred to as ‘non-breeding’), to represent an average prevalence across the population35. We report prevalence by sample type tested, as prevalenceblood (refers to population prevalence in blood samples) and prevalencecloacal (refers to population prevalence in cloacal swabs).
We analysed BFDV prevalence and host body mass using generalized linear models (GLM), which are commonly used as a flexible method for analysis of seasonal variation64. For binary data, we report Binomial (Clopper-Pearson) ‘exact’ CI as a widely used, conservative method recommended for small sample sizes65. We used a binomial distribution with a logit link to analyse effects on prevalence and a Gaussian distribution with identity link for the analysis of body condition and body mass. Where applicable, we checked the residuals to confirm that they conformed to the model assumptions. We report model outputs in Supplementary Tables S3–S11. In all figures and the main body of the text, we show raw data, i.e. means with 95% confidence intervals (CI). To report model fit values for binary data (e.g. infection status) we used the Nagelkerke R2. For continuous data (e.g. body mass), we show the overall R2 created by univariate analysis of variance.
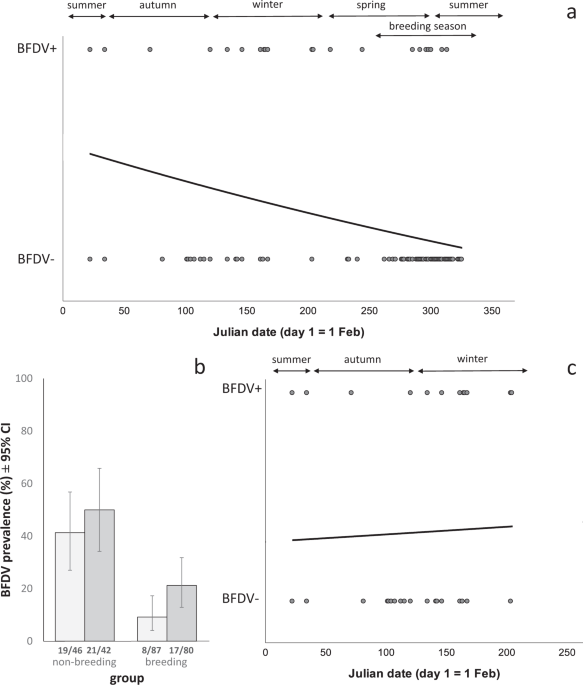
We analysed temporal changes in BFDV prevalence using two predictors: season (spring, summer, autumn, winter) and Julian date (‘date’; with the 1st February set as day 1). We analysed date as a continuous variable in addition to season, because grouping samples by season can result in samples that were in fact collected close together being assigned to different seasons, potentially leading to unreliable results (for example, a bird trapped on the 31st May would be in the autumn group, but a bird trapped one day later would be in the winter group). Both season (as a categorical predictor) and date (as a continuous predictor) are common methods for investigating seasonal effects64,66. Seasons were defined as follows: Spring: 1st September to 30th November; summer: 1st December to 28th February; autumn: 1st March to 31st May; winter: 1st June to 31st August. Season was included as an ordinal variable (season 1 = spring, 2 = summer, 3 = autumn, 4 = winter). During exploratory data analysis we ran the same models using season as either ordinal or nominal variable, which led to identical results. Julian date was set to day one on the 1st February, to represent a biologically significant date67, i.e. the end of the breeding season of P. elegans in our study area35. During exploratory data analysis, we also tested 1st January (start of the year) and 1st September (beginning of the breeding season) as Julian day 1. Between the three examples of Julian day 1, the resulting R2, AICc and p-values were very similar (Supplementary Table S11); we therefore only report results based on 1st February as day 1. We centred Julian date to avoid collinearity, and tested linear, quadratic and cubic date predictors. For blood samples, models with quadratic date terms showed better fit (based on AICc and Nagelkerke R2) than models with cubic as well as quadratic date terms (data not shown). For body condition and body mass, models had almost identical fit with and without the cubic date terms. We therefore chose to use the most parsimonious models and for blood samples, body condition and body mass, we report model outputs without the cubic date term. For cloacal swabs and when testing our complete data set of P. elegans, models including cubic as well as quadratic date terms showed better fit than models that included only quadratic terms. We thus report results based on these models (Supplementary Table S4). Additional predictors we used were host traits that have previously been shown to influence BFDV infection (age class and sex)56. After performing GLMs on the full data set, we also tested seasonal effects separately in subsets of breeding and non-breeding birds, as date and breeding status were correlated. Nine birds which were caught in walk-in traps during the breeding season were excluded from analyses comparing breeding and non-breeding birds, as we could not determine whether or not they were breeding at the time of capture. Sample size thus varies between models, and also because of missing information for some predictors for some individual birds.
We repeated models analysing body mass by including tarsus length as a predictor, to estimate body condition by accounting for individual size differences. Body condition results are only shown in Supplementary Table S5, as they are very similar to results with body mass which did not include tarsus length (Supplementary Table S6). To compare viral load between groups (males vs females, breeding vs non-breeding) we conducted t-tests35.
Source: Ecology - nature.com



