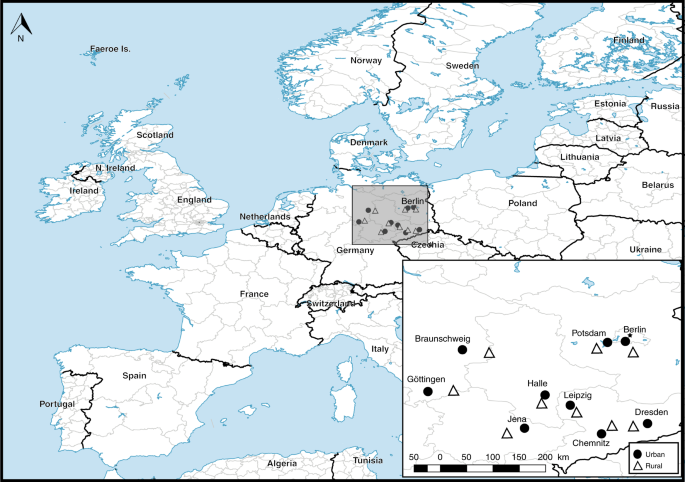
Study sites
To test the association between urban versus rural land use on insect pollinators and pollination, we used a well-replicated study design that employed a flower-rich urban site paired with a flower-rich rural site, replicated across nine major cities in central and eastern Germany. Cities were Berlin, Braunschweig, Chemnitz, Dresden, Göttingen, Halle, Jena, Leipzig and Potsdam (Fig. 1). The minimum distance between two city sites was 20 km (Fig. 1). Each adjacent rural site was a minimum of 10 km (maximum 40 km, mean 25 km; see Fig. 1) from its urban paired site and greater than 25 km from all other rural or urban sites (Fig. 1), sufficient distance for all 18 sites to be considered independent. All sites were selected to represent flower-rich habitats within the urban and rural landscapes of Central Europe (Supplementary Table 1).
Due to the well-documented relationship between pollinator diversity and flower richness49, we chose our sites to reflect a flower-rich and pollinator-optimal comparison of the two ecosystems (Supplementary Table 1), allowing us to focus on the broader landscape context (i.e. urban versus rural ecosystems). Our logic was to compare the best urban sites for insect pollinators15,22 with the best rural sites that were matched in terms of habitat structure (land cover, flower abundance), though each pair was sited in a different land use matrix (urban versus rural, respectively). At each site (urban and rural), we selected a 25 m × 25 m area with diverse floral resources, which we used as our urban sampling plot (Supplementary Table 1). Though sampling across a gradient of urbanisation and into the rural landscape is another sampling strategy that has been used to demonstrate the importance of cities for pollinators19,20,22, our intention was to compare urban with rural habitats and thus we maximised the number of urban–rural comparisons for our given intensity of sampling.
Urban sites were near the urban core, surrounded by high road density and human infrastructure, and were located in or in close proximity to botanical gardens (N = 7) or public parks (N = 2). They differed in their surrounding urban matrix, though residential and commercial/industrial areas comprised a mean of 60% of the surrounding (1000 m scale) land use across all urban sites (Supplementary Table 6). Private gardens were not used for experiments due to lack of availability at all sites and access restrictions. At each site, we selected a 25 m × 25 m area with diverse floral resources, which we used as our urban sampling plot (Supplementary Table 1).
Rural sites were selected to be as similar as urban sites in terms of their local (250 m scale) land cover (i.e. flower abundance) by using land cover maps within a Geographic Information System (Quantum GIS). Arable land (=agricultural land) and forest/semi-natural cover were the dominant land use types at the landscape scale, comprising a mean of 45% and 41% the of surrounding (1000 m scale) land use across all rural sites, respectively (Supplementary Table 7), typical of the region’s rural environment i.e. our rural sites were not impacted by urban sprawl. To identify rural sites that were ideal for insect pollinators yet independent of urban sites, we drew a buffer at a circumference of 10 km radius from each urban site and then used GIS to identify areas of semi-natural grassland and scrub vegetation immediately outside the buffer that were largely devoid of ‘residential’ and ‘commercial/industrial’ and dominated by arable land and forest/semi-natural cover within the surrounding 1 km radius. Candidate rural sites were then visited and one was randomly selected that had a 25 m × 25 m area near its geographical centre with diverse floral resources (Supplementary Table 1), which we used as our rural sampling plot. By using these criteria for site selection, we aimed to sample from the best sites for insect pollinators, and potentially also for pollination, among urban and among rural localities.
Measurement of flying insect diversity
To compare the diversity of flying insects at urban flower-rich sites with those at rural flower-rich sites, we measured species richness of the four main orders of flying insect pollinators: Diptera, Lepidoptera, Coleoptera, and Hymenoptera50. Though our experimental Trifolium pratense plants were primarily visited by bees (see above), our attention to all four orders allows us to address the pollinators of a broader range of plant species, and to explore their taxonomic diversity in response to urbanisation. We employed pan traps to sample insects, which are a standardised and commonly used method for collecting flying insects that have been widely used in studies of pollinator communities51,52. As for any other flying insect sampling method, pan traps have disadvantages such as potential taxonomic bias and under-sampling of large insects. However, in a comparative study, they were found to be the most efficient method of sampling bees across geographic regions in Europe51. Additionally, coloured pan traps have been found to outperform malaise traps in sampling all major insect pollinators, including Diptera, Lepidoptera and Coleoptera52.
Each urban–rural site pair was visited at the same time and for a total of five consecutive warm, non-windy days at one point between June and August 2014 (Supplementary Table 8). Temperatures exceeded 16 °C, wind speed was less than 2 m/s at 1 m above ground level, and skies were sunny (<50% cloud cover; Supplementary Table 8) on all sampling days. Insects were sampled using three blue, three yellow and three white pan traps (diameter: 42 cm, height: 2.8 cm) mounted on a stick at vegetation height at each site and set out across the 25 m × 25 m plot. Each pan trap was 2/3 filled with unscented soapy water and emptied every day. Insects from traps were stored in 95% ethanol. Insect samples from each site were washed, dried and weighed using a balance (Denver Instruments SI-2002A, Denver, USA).
For assessment of species richness, we used next generation sequencing (NGS)-based metabarcoding53 to identify the number of different species of Diptera, Lepidoptera, Coleoptera, and Hymenoptera based on mitochondrial COI DNA sequences. Metabarcoding has been successfully used to assess patterns of arthropod diversity, and has proven faster, cheaper and more efficient than traditional morphological taxonomy53,54. This is likely to be particularly the case for the large and diverse order Diptera55. A disadvantage of this approach is that it provides only a list of species sampled at a site, not their relative abundance, and it may have under-recorded small or rare species that occurred as singletons in our pan trap material. Our metabarcoding protocol combined mass-PCR amplification of the COI barcode gene, 454-pyrosequencing and several optimised bioinformatics steps to determine OTUs and perform taxonomic assignment to species (Supplementary Methods).
In brief, denoised 454-pyrosequencing reads of pan-trap material from all 18 sites were reduced 1.8-fold from 157,310 raw reads to 85,223 Insecta-only sequences (orders: Diptera, Lepidoptera, Coleoptera and Hymenoptera). After removal of singletons, our total metabarcoding dataset contained 592 Insecta OTUs. Of these, 342 (57.8%) belonged to Diptera, 116 (19.6%) to Hymenoptera, 81 (13.7%) to Lepidoptera and 53 (9.0%) to Coleoptera (the main insect orders sampled; see Supplementary Fig. 4). The majority of OTUs (308 out of 592, 66.2%) were successfully assigned to species level (Supplementary Dataset). For Diptera we could assign 117 OTUs (34.2%); for Hymenoptera 85 OTUs (73.2%); for Lepidoptera 65 OTUs (80.2%) and for Coleoptera 41 OTUs (77.3%) to species (Supplementary Dataset), a rate which is typical for Central European barcoding studies55. We henceforth use species richness and OTU richness interchangeably. The number of reads was not correlated with OTU richness across our dataset (Pearson’s product-moment correlation r = 0.033, N = 18, P > 0.05), suggesting we had sufficient reads to saturate our OTU assessment per site.
To explore whether phylogenetic diversity and, by inference, trait diversity56 of flying insects were related to the ecosystem service of pollination and how they were impacted by urbanisation, we additionally estimated two phylogenetic diversity metrics, (i) PSV and (ii) MNTD, using the R package picante57. PSV quantifies how phylogenetic relatedness decreases the variance of a hypothetical unselected/neutral trait shared by all species in a community. The expected value of PSV is statistically independent of species richness; it is 1 when all species in a sample are unrelated and approaches 0 as species become more related58. MNTD provides an average of the distances between each species and its nearest phylogenetic neighbour in the community. MNTD quantifies the degree that a community may be a set of closely related species versus a heterogeneous set of taxa from disparate taxonomic clades59. It was only weakly related to PSV in our dataset (Pearson’s product-moment correlation r = 0.441, N = 18, P = 0.066) and therefore we retained it in analyses as a second measure of phylogenetic diversity.
Quantifying the ecosystem service of pollination
To quantify differences in pollination service provision in urban versus rural ecosystems, we used greenhouse-raised Trifolium pratense plants as ‘pollinometers’ at each site and evaluated their pollination success. Though T. pratense is preferentially visited by long-tongued bumble bees22,60, in a previous study22,41 we found high correlations in the pollination success of Trifolium pratense, Borago officinalis, Sinapis alba and Trifolium repens experimental plants (common flowering plants in Germany; results in Supplementary Fig. 5 and Supplementary Table 9), suggesting that T. pratense could be used as a model system to quantify the ecosystem service of pollination in our study region.
Seeds of wild T. pratense were obtained from a local seed provider (Rieger-Hofmann GmbH, Blaufelden, Germany). These (diploid) wild plants were readily visited by bumble bees (Bombus spp.), honey bees (Apis mellifera) and other wild bees (other Anthophila) (see results). All seeds were germinated and grown for two months in an insect-free glasshouse before placement at our study sites. Ten potted plants, each with eight open inflorescences (flower heads) (N = 8 per plant) marked with coloured tape, were placed at each field site during the five flying insect sampling days at that site. In each plant, one inflorescence (entire flower head containing ca. 100 individual flowers) of the eight was bagged throughout the experiment in the field with fine net (1 mm gauze) to prevent visitation by pollinators (zero control). Bagged flowers did not set any seed, demonstrating the dependence of T. pratense on insect visitation for seed set.
Pollinometer plants in each community were randomly ordered at one metre distance along a transect of 10 m × 1 m within the 25 m × 25 m plot at each site. Once the 5 days of the pollination experiment were completed at a site, focal plants were returned to the insect-free greenhouse until seeds were formed. Seeds from all seven unbagged inflorescences per plant were counted and the average number of seeds per plant (i.e. per seven inflorescences) was used as a measure of the ecosystem service of pollination. Each inflorescence of T. pratense contains up to 100 flowers. We weighed all marked, dried inflorescences per plant as a surrogate of the number of flowers in each inflorescence and tested for differences in flower number per inflorescence between the two ecosystems. We did not find any differences in the weight of inflorescences between the two ecosystems (LMM, t = 0.183, P = 0.855), suggesting that an equivalent number of flowers had been exposed to pollinators at each member of a site pair. Thus, our method of assessment of the ecosystem service of pollination did not differ between an urban-rural pair of sites and is presented in terms of number of seeds per plant (i.e. per seven inflorescences per plant exposed to pollinators).
In addition, we monitored all insects visiting the flowers of experimental T. pratense plants for five hours at each site in order to estimate flower visitation rates because these are causally related to pollination success. Due to the possible confounding effect of flower visitation frequency when testing the effect of pollinator diversity on pollination61, we used flower visitation rate as a covariate in our statistical analysis of the relationship between pollinator diversity and pollination. Each experimental plant was observed twice per site (15 min in the morning and 15 min in the afternoon), for a total of 300 min observation time of T. pratense pollinometers per site. Visitor identity (11 morphogroups: Coleoptera; Syrphidae; other Diptera; Lepidoptera; wasps; bees of each family: Andrenidae, Halictidae; plus each morphospecies group: Bombus lapidarius/Bombus sorooensis proteus; Bombus terrestris/Bombus lucorum; Bombus pascuorum; and Apis mellifera) was recorded. Furthermore, at each site we estimated the abundance of conspecific pollen donors by counting the number of inflorescences of co-flowering T. pratense plants within a 200 m buffer around each plot (Supplementary Table 1).
Local patch and landscape variables
To determine the main ecological correlates of insect biodiversity and pollination in both rural and urban flower-rich sites, we gathered a series of local (patch) and landscape-scale variables potentially related to insect pollinators and pollination. Although we selected sites with high availability of floral resources in both ecosystems, we additionally quantified local flowering plant richness and abundance (number of flowers of each plant species) as an estimator of floral resource availability using 10 randomly placed 1 m2 quadrats at each of our sampling sites (Supplementary Table 1). These estimates were used in our models as covariates. We also quantified habitat composition at five spatial scales (250, 500, 750, 1000 and 1500 m) per site using Quantum GIS and land cover data obtained from Geofabrik GmbH (http://www.geofabrik.de/). We calculated landscape diversity for each radius using the Shannon–Wiener index (Hs): Hs = −∑pi • ln pi, where pi is the proportion of each land cover of type i, as defined in Supplementary Table 10.
To identify the scale at which surrounding land cover had the most power to explain each insect order’s occurrence, we correlated each insect order’s OTU richness with landscape diversity (measured as Shannon–Weiner diversity of proportions of land use types) at each of our study sites at all five scales62. Correlation coefficients peaked at the 250 m scale for Diptera and Lepidoptera OTU richness and at the 1000 m scale for Coleoptera and Hymenoptera OTU richness both across (combining urban and rural sites in one analysis) and within each ecosystem (analysing urban and rural sites separately) (Supplementary Table 11). These spatial scales for each taxon were then used for subsequent landscape-scale analyses.
Several metrics known to impact flying insect biodiversity were used to quantify landscape heterogeneity (composition and configuration) at each of the 18 sites at both 250 and 1000 m scales28,63. These were (i) the proportion of semi-natural cover (grassland, meadows and scrub vegetation), (ii) the proportion of forest, (iii) the extent of arable (=agricultural) cover, (iv) the proportion of residential and (v) commercial/industrial land cover, (vi) the extent of botanical gardens, public parks and allotments, and (vii) edge density, as total length of ‘green cover’ (semi-natural and forest cover, botanical gardens, public parks, and allotments) patch edges divided by the total area, and which represents a quantification of landscape configuration.
Statistical analyses
Given our paired ‘urban-rural’ experimental design, the rationale in our statistical analyses was to use site pair as a random factor and to compare between ecosystem type (urban versus rural) because sites had been selected a priori as belonging to either urban or rural ecosystems. We controlled for potentially confounding local and landscape factors, unless we specifically aimed to model their relationship to predictor variables: dimensions of biodiversity and pollination. Results reported below were qualitatively the same in all analyses when we did not control for potentially confounding local and landscape variables, supporting the view that we had selected equivalent sites in urban and rural ecosystems and reinforcing the robustness of our results.
To derive comparable estimates across urban and rural sites, we standardized all quantitative predictors to a mean of zero and standard deviation of one. Prior to each analysis we used variance inflation factors (VIFs) to check for collinearity among our explanatory variables. VIFs were lower than 3 for all predictors in all models tested, indicating negligible levels of collinearity64.
To determine whether flower-rich urban sites supported a higher insect pollinator biodiversity than flower-rich rural sites, we tested for differences in flying insect species richness between rural and urban sites using generalised linear mixed models (GLMMs) with Poisson error structure and site pair as a random effect factor. We undertook these analyses for all insects and for each insect order separately, using all local patch (i.e. flower richness and abundance) and landscape variables (i.e. proportion of semi-natural cover, forest, arable, residential, commercial/industrial, botanical gardens/parks/allotments, habitat diversity and edge density) as covariates. For the Diptera and Hymenoptera datasets, we additionally analysed subsets of each, namely the hover flies (Diptera: Syrphidae) and the bees (Hymenoptera: Anthophila), as they are considered as the most important insect pollinator taxa within their respective orders50. We also tested for differences in flying insect biomass (log of weight in g) between urban and rural sites using a linear mixed model (LMM). To test for differences between rural and urban flower-rich sites with respect to our phylogenetic diversity metrics, we used LMMs for MNTD and GLMMs with binomial error structure for PSV. Again, site pair was included as a random effect factor and all local patch and landscape-scale factors described above were included as covariates.
Flower-rich sites located within an inhospitable landscape may attract insects from further afield than sites nested within a floristically rich landscape, a ‘honeypot’ effect. Though we did not quantify floral abundance across the wider landscape so as to test explicitly for the honeypot effect, we tested for differences between urban versus rural sites in terms of local (flower abundance, flower diversity) and landscape variables (green cover, edge density) using LMMs and GLMMs to assess whether they varied consistently between ecosystems.
To analyse the effects of local (patch) habitat (i.e. flower richness and abundance) and landscape composition (i.e. proportion of semi-natural cover, forest, arable, residential, commercial/industrial and botanical gardens/parks/allotments and habitat diversity) and configuration (i.e. edge density) on species diversity for each insect order, we used generalised linear models (GLMs) with Julian day as a covariate. We explored these drivers of insect diversity for rural and urban ecosystems separately because sites had been selected a priori to represent flower-rich locations typical of each ecosystem but with stark differences between ecosystem types (urban: mean 60% residential/industrial land cover; rural: mean 86% agricultural/forest/semi-natural land cover). For each insect order, we used the spatial scale derived from their response to landscape diversity (for Diptera and Lepidoptera, 250 m; for Coleoptera and Hymenoptera, 1000 m, see the “Results” section) and Julian day was used as a covariate.
To test for differences in insect community composition between flower-rich urban and rural sites, we performed a paired permutational multivariate analysis of variance using the adonis function, with 999 permutations, implemented in the R package vegan65. In the adonis analysis, the Jaccard distance matrix of species composition was the response variable, with ecosystem (urban/rural) as a fixed factor. The strata (block) argument was set to ‘site pair’ so that randomizations were constrained to occur within each pair and not across all sample sites. We undertook these analyses for all insects and for Hymenoptera only. We employed non-metric multidimensional scaling (NMDS) within the package vegan to visualize the variation in community composition. For each site we also calculated the mean ecological distance (Jaccard index) over all pairwise comparisons of the 9 sites belonging to the same ecosystem type. We used a LMM to compare urban and rural ecosystems, with pair as a random effect factor and using all local patch and landscape variables as covariates.
We tested the effects of each ecosystem (urban versus rural) on the pollination of T. pratense plants using LMMs. In doing so, we included local flower richness, local flower abundance, abundance of co-flowering T. pratense plants and landscape-scale factors (i.e. proportion of semi-natural cover, forest, arable, botanical gardens/public parks/allotments, habitat diversity and edge density) as covariates. Individual plants were nested within each site and site pair was used as a random effect.
To explore the main environmental correlates of visitation rates in each ecosystem (urban, rural), we used GLMs, exactly as we did when testing the main environmental correlates of insect species diversity (i.e. for each ecosystem separately).
We then used LMMs to explore the relationships between insect biodiversity and T. pratense seed set while controlling for flower visitation rates. In doing so, site pair was used as a random effect and ecosystem (urban versus rural) as a fixed effect factor. We modelled seed set as dependent on insect species richness, PSV and MNTD, measured over all Insecta and for each insect order separately.
When investigating the main environmental drivers of insect species diversity, we used an all-subset (i.e. all combinations of predictors of interest) automated model selection approach based on the Akaike Information Criterion, corrected for small sample size (AICc), using the dredge function (R package MuMIn66) and with a maximum of three predictors to avoid model overfitting. All mixed model analyses were performed using the R package lme467. All model (GLMMs, LMMs and GLMs) assumptions were checked visually. The residuals of all regression models were tested for spatial and temporal autocorrelation using Moran’s I implemented in the R package ape68. Residuals were not found to be autocorrelated (P > 0.05).
Finally, having identified differences in pollinator diversity and pollination service provision at urban versus rural sites, and the local and landscape factors potentially driving urban-rural differences, we synthesised our analyses by exploring commonalities across all (urban and rural) sites. Here our aim was to identify the most important putative causes of variation in pollination, regardless of ecosystem type, and hence we did not use ecosystem type (urban/rural) as a fixed factor in analyses, though we did retain pair as a random effect factor to account for our experimental design in which pairs of sites were investigated sequentially across the summer. To do so, we used piecewise SEM, which allowed us to visualise and also statistically test for the importance of factors and their interrelations in a logical, causal path. In keeping with the main rationale of our study, we also used SEMs to identify the main factors associated with pollinator biodiversity and pollination in rural versus urban sites by running separate SEMs for each ecosystem. The similarity of SEMs for each ecosystem (Supplementary Tables 8a and b), dominated by the predictors: edge density, Hymenoptera species richness and Hymenoptera phylogenetic diversity as well as plant visitation rate, justify our synthetic approach of exploring factors affecting pollination across both ecosystems within one SEM.
In generating all SEMs, we hypothesised that landscape composition and configuration as well as local patch flower richness and abundance, and conspecific pollen donor availability might have affected T. pratense seed set directly and also indirectly through affecting insect visitation rates, OTU richness and phylogenetic diversity (PSV and MNTD; data for Diptera, Lepidoptera, Coleoptera and Hymenoptera added separately). We performed piecewise SEM analyses using the R package piecewiseSEM69. From an overall model based on a priori knowledge of interactions with all hypothesised effects, we used a backwards stepwise elimination process based on AICc to remove non-significant pathways. In addition, we used the d-separation (d-sep) test to evaluate whether the non-hypothesised, independent paths were significant and whether the models incorporated into the SEM could be improved with the inclusion of any of the missing path(s). We assessed goodness of fit of the final model using the Fisher’s C statistic. Path coefficients and deviance explained were then calculated for each model. We report both conditional (R2c, all factors) and marginal (R2m, fixed factors only) coefficients of determination for generalized linear mixed effect models incorporated in the SEM.
All statistical analyses were performed in R v. 3.5.270.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Source: Ecology - nature.com



