Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103:3846–51.
Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, Parkes RJ, et al. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433:861–4.
Lipp JS, Morono Y, Inagaki F, Hinrichs K-U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature. 2008;454:991–4.
Buongiorno J, Turner S, Webster G, Asai M, Shumaker AK, Roy T, et al. Interlaboratory quantification of Bacteria and Archaea in deeply buried sediments of the Baltic Sea (IODP Expedition 347). FEMS Microbiol Ecol. 2017;93:fix007.
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci USA. 2008;105:10583–8.
Youssef NH, Rinke C, Stepanauskas R, Farag I, Woyke T, Elshahed MS. Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum “Diapherotrites. ISME J. 2015;9:447–60.
Farag IF, Youssef NH, Elshahed MS. Global distribution patterns and pangenomic diversity of the candidate phylum “Latescibacteria” (WS3). Appl Environ Microbiol. 2017;83:e00521–17.
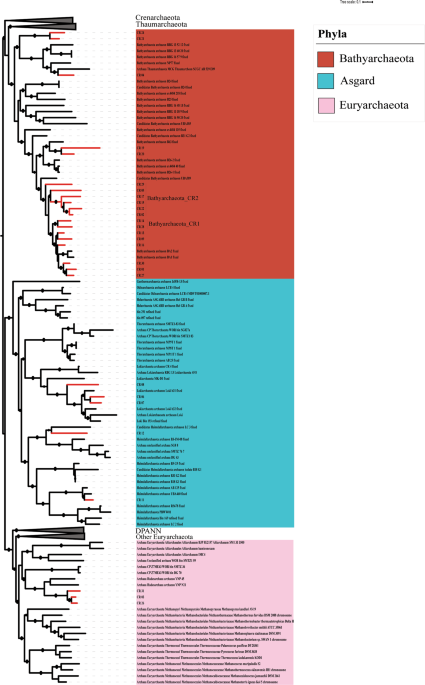
Kubo K, Lloyd KG, Biddle F, Amann J, Teske R, Knittel A, et al. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 2012;6:1949–65.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7.
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–9.
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048.
Castelle CJ, Banfield JF. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 2018;172:1181–97.
Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219.
Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8.
Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, et al. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol. 2019;4:1138–48.
MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol. 2019;5:48–61.
Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, et al. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun. 2019;10:1822.
Zhou Z, Pan J, Wang F, Gu J-D, Li M. Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol Rev. 2018;42:639–55.
Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–8.
He Y, Li M, Perumal V, Feng X, Fang J, Xie J, et al. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol. 2016;1:16035.
Martino A, Rhodes ME, León-Zayas R, Valente IE, Biddle JF, House CH. Microbial diversity in sub-seafloor sediments from the Costa Rica margin. Geosciences. 2019;9:218.
Vannucchi P, Ujiie K, Stroncik N, IODP Exp. 334 Scientific Party, Yatheesh V. IODP expedition 334: An investigation of the sedimentary record, fluid flow and state of stress on top of the seismogenic zone of an erosive subduction margin. Sci Drill. 2013;15:23–30.
Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8.
Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome. 2014;2:26.
Laczny CC, Sternal T, Plugaru V, Gawron P, Atashpendar A, Margossian HH, et al. VizBin—an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome. 2015;3:1.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8.
Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119.
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). New Orleans, LA: Institute of Electrical and Electronics Engineers; 2010. p. 1–8. https://ieeexplore.ieee.org/abstract/document/5676129.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010;11:431.
Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31.
Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–51.
Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–3.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–32.
Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–9.
Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40.
Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8:R16.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Mendler K, Chen H, Parks DH, Lobb B, Hug LA, Doxey AC. AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 2019;47:4442–8.
Stagars MH, Ruff SE, Amann R, Knittel K. High diversity of anaerobic alkane-degrading microbial communities in marine seep sediments based on (1-methylalkyl)succinate synthase genes. Front Microbiol. 2015;6:1511.
Flamholz A, Noor E, Bar-Even A, Milo R. eQuilibrator-the biochemical thermodynamics calculator. Nucleic Acids Res. 2012;40:D770–5.
Lever MA. Acetogenesis in the energy-starved deep biosphere—a paradox? Front Microbiol. 2011;2:284.
LaRowe DE, Amend JP. Catabolic rates, population sizes and doubling/replacement times of microorganisms in natural settings. Am J Sci. 2015;315:167–203.
Dick JM. Calculation of the relative metastabilities of proteins using the CHNOSZ software package. Geochem Trans. 2008;9:10.
Helgeson HC. Thermodynamics of hydrothermal systems at elevated temperatures and pressures. Am J Sci. 1969;267:729–804.
Levin LA, Orphan VJ, Rouse GW, Rathburn AE, Ussler W, Cook GS, et al. A hydrothermal seep on the Costa Rica margin: middle ground in a continuum of reducing ecosystems. Proc Biol Sci. 2012;279:2580–8.
Meckenstock RU, Boll M, Mouttaki H, Koelschbach JS, Cunha Tarouco P, Weyrauch P, et al. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. J Mol Microbiol Biotechnol. 2016;26:92–118.
Song B, Ward BB. Genetic diversity of benzoyl coenzyme A reductase genes detected in denitrifying isolates and estuarine sediment communities. Appl Environ Microbiol. 2005;71:2036–45.
López Barragán MJ, Carmona M, Zamarro MT, Thiele B, Boll M, Fuchs G, et al. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J Bacteriol. 2004;186:5762–74.
Huang J-M, Baker BJ, Li J-T, Wang Y. New microbial lineages capable of carbon fixation and nutrient cycling in deep-sea sediments of the Northern South China Sea. Appl Environ Microbiol. 2019;85:e00523–19.
Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016;10:761–77.
Kouzuma A, Kato S, Watanabe K. Microbial interspecies interactions: recent findings in syntrophic consortia. Front Microbiol. 2015;6:477.
Pirbadian S, Barchinger SE, Leung KM, Byun HS, Jangir Y, Bouhenni RA, et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci USA. 2014;111:12883–8.
Dong X, Greening C, Rattray JE, Chakraborty A, Chuvochina M, Mayumi D, et al. Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat Commun. 2019;10:1816.
Glombitza C, Jaussi M, Røy H, Seidenkrantz M-S, Lomstein BA, Jørgensen BB. Formate, acetate, and propionate as substrates for sulfate reduction in sub-arctic sediments of Southwest Greenland. Front Microbiol. 2015;6:846.
McCrindle SL, Kappler U, McEwan AG. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv Microb Physiol. 2005;50:147–98.
Evans PN, Boyd JA, Leu AO, Woodcroft BJ, Parks DH, Hugenholtz P, et al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17:219–32.
Adam PS, Borrel G, Gribaldo S. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc Natl Acad Sci USA. 2018;115:E1166–73.
Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, et al. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome. 2013;1:22.
Mehrshad M, Salcher MM, Okazaki Y, Nakano S-I, Šimek K, Andrei A-S, et al. Hidden in plain sight-highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome. 2018;6:176.
Jørgensen BB, Boetius A. Feast and famine-microbial life in the deep-sea bed. Nat Rev Microbiol. 2007;5:770–81.
Liu Y-F, Qi Z-Z, Shou L-B, Liu J-F, Yang S-Z, Gu J-D, et al. Anaerobic hydrocarbon degradation in candidate phylum “Atribacteria” (JS1) inferred from genomics. ISME J. 2019;13:2377–90.
Mausz MA, Chen Y. Microbiology and ecology of methylated amine metabolism in marine ecosystems. Curr Issues Mol Biol. 2019;33:133–48.
Source: Ecology - nature.com



