Shah Walter, S. R. et al. Microbial decomposition of marine dissolved organic matter in cool oceanic crust. Nat. Geosci. 11, 334–339 (2018).
D’Hondt, S., Rutherford, S. & Spivack, A. J. Metabolic activity of subsurface life in deep-sea sediments. Science 295, 2067–2070 (2002).
Jørgensen, B. B. Deep subseafloor microbial cells on physiological standby. Proc. Natl Acad. Sci. USA 108, 18193–18194 (2011).
Hoehler, T. M. & Jørgensen, B. B. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94 (2013).
Tully, B. J., Wheat, C. G., Glazer, B. T. & Huber, J. A. A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 12, 1–16 (2018).
Santelli, C. M., Edgcomb, V. P., Bach, W. & Edwards, K. J. The diversity and abundance of bacteria inhabiting seafloor lavas positively correlate with rock alteration. Environ. Microbiol. 11, 86–98 (2009).
Jungbluth, S. P., Bowers, R. M., Lin, H. T., Cowen, J. P. & Rappé, M. S. Novel microbial assemblages inhabiting crustal fluids within mid-ocean ridge flank subsurface basalt. ISME J. 10, 2033–2047 (2016).
Shrenk, M. O., Huber, J. A. & Edwards, K. J. Microbial provinces in the subseafloor. Ann. Rev. Mar. Sci. 2, 279–304 (2010).
Mason, O. U. et al. First investigation of the microbiology of the deepest layer of ocean crust. PLoS ONE 5, e15399 (2010).
Zhang, X., Feng, X. & Wang, F. Diversity and metabolic potentials of subsurface crustal microorganisms from the western flank of the Mid-Atlantic Ridge. Front. Microbiol. 7, 363 (2016).
Früh-Green, G. L. et al. Magmatism, serpentinization and life: insights through drilling the Atlantis Massif (IODP Expedition 357). Lithos 323, 137–155 (2018).
Lipp, J. S. & Hinrichs, K.-U. Structural diversity and fate of intact polar lipids in marine sediments. Geochim. Cosmochim. Acta 73, 6816–6833 (2009).
Valentine, D. L. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5, 316–323 (2007).
Summons, R. E. & Lincoln, S. A. in Fundamentals of Geobiology (eds Knoll, A. H.) 269–296 (John Wiley and Sons, 2012).
Swan, B. K. et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300 (2011).
Sheik, C. S., Jain, S. & Dick, G. J. Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317 (2014).
Grossi, V. et al. Mono- and dialkyl glycerol ether lipids in anaerobic bacteria: biosynthetic insights from the mesophilic sulfate reducer Desulfatibacillum alkenivorans PF2803T. Appl. Environ. Microbiol. 81, 3157–3168 (2015).
Hawley, A. K. et al. Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 8, 1507 (2017).
Kelley, D. S. et al. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307, 1428–1434 (2005).
Puente-Sánchez, F. et al. Viable cyanobacteria in the deep continental subsurface. Proc. Natl Acad. Sci. USA 115, 10702–10707 (2018).
Klein, F., Grozeva, N. G. & Seewald, J. S. Abiotic methane synthesis and serpentinization in olivine-hosted fluid inclusions. Proc. Natl Acad. Sci. USA 116, 17666–17672 (2019).
Zolotov, M. & Shock, E. L. Abiotic synthesis of polycyclic aromatic hydrocarbons on Mars. J. Geophys. Res. Planets 104, 14033–14049 (1999).
Fonknechten, N. et al. Clostridium sticklandii, a specialist in amino acid degradation:revisiting its metabolism through its genome sequence. BMC Genomics 11, 555 (2010).
Cai, L. et al. Comparative genomics study of polyhydroxyalkanoates (PHA) and ectoine relevant genes from Halomonas sp. TD01 revealed extensive horizontal gene transfer events and co-evolutionary relationships. Microb. Cell Fact. 10, 88 (2011).
Jendrossek, D. & Handrick, R. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56, 403–432 (2002).
Liu, G. et al. Enoyl-CoA hydratase mediates polyhydroxyalkanoate mobilization in Haloferax mediterranei. Sci. Rep. 6, 24015 (2016).
Han, J. et al. Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) producer. J. Bacteriol. 194, 4463–4464 (2012).
Lin, H.-T. et al. Inorganic chemistry, gas compositions and dissolved organic carbon in fluids from sedimented young basaltic crust on the Juan de Fuca Ridge flanks. Geochim. Cosmochim. Acta 85, 213–227 (2012).
Santos-Beneit, F. The Pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6, 402 (2015).
Zinke, L. A. et al. Thriving or surviving? Evaluating active microbial guilds in Baltic Sea sediment. Environ. Microbiol. Rep. 9, 528–536 (2017).
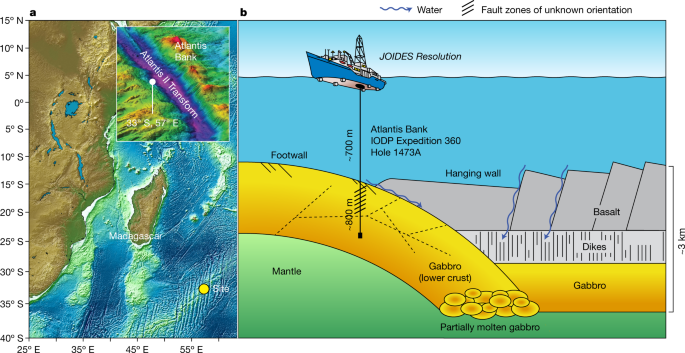
Dick, H. J. B. et al. The Atlantis Bank gabbro massif, Southwest Indian Ridge. Prog. Earth Planet. Sci. 6, 64 (2019).
Fox, P. J. & Gallo, D. G. A tectonic model for ridge-transform-ridge plate boundaries: implications for the structure of oceanic lithosphere. Tectonophysics 104, 205–242 (1984).
Dick, H. J. B. et al. Dynamic accretion beneath a slow-spreading ridge segment: IODP hole 1473A and the Atlantis Bank oceanic core complex. J. Geophys. Res. Solid Earth 124, 12631–12659 (2019).
Baines, A. G. et al. Mechanism for generating the anomalous uplift of oceanic core complexes: Atlantis Bank, southwest Indian Ridge. Geology 31, 1105–1108 (2003).
Morono, Y., Terada, T., Kallmeyer, J. & Inagaki, F. An improved cell separation technique for marine subsurface sediments: applications for high-throughput analysis using flow cytometry and cell sorting. Environ. Microbiol. 15, 2841–2849 (2013).
Lundin, A., Hasenson, M., Persson, J. & Pousette, A. Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol. 133, 27–42 (1986).
Coolen, M. J. & Overmann, J. Functional exoenzymes as indicators of metabolically active bacteria in 124,000-year-old sapropel layers of the eastern Mediterranean Sea. Appl. Environ. Microbiol. 66, 2589–2598 (2000).
Pella, E. Elemental organic analysis. Part 1, historical developments. Am. Lab. 22, 116–125 (1990).
Pella, E. Elemental organic analysis. Part 2: State of the art. Am. Lab. 22, 28–32 (1990).
Whiteside, J. H. et al. Pangean great lake paleoecology on the cusp of the end-Triassic extinction. Palaeogeogr. Palaeoclimatol. Palaeoecol. 301, 1–17 (2011).
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M. & Hinrichs, K. U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18, 617–628 (2004).
Klein, A. T. et al. Investigation of the chemical interface in the soybean–aphid and rice–bacteria interactions using MALDI-mass spectrometry imaging. Anal. Chem. 87, 5294–5301 (2015).
Becker, K. W. et al. An improved method for the analysis of archaeal and bacterial ether core lipids. Org. Geochem. 61, 34–44 (2013).
Parada, A. E., Needham, D. M. & Fuhrman, J. A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Sheik, C. S. et al. Identification and removal of contaminant sequences from ribosomal gene databases: lessons from the Census of Deep Life. Front. Microbiol. 9, 840 (2018).
Cole, J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 (2009).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016).
Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007).
Glassing, A., Dowd, S. E., Galandiuk, S., Davis, B. & Chiodini, R. J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 8, 24 (2016).
Le Calvez, T., Burgaud, G., Mahé, S., Barbier, G. & Vandenkoornhuyse, P. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 75, 6415–6421 (2009).
Burgaud, G., Arzur, D., Durand, L., Cambon-Bonavita, M.-A. & Barbier, G. Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol. Ecol. 73, 121–133 (2010).
Valentine, D. L. et al. Propane respiration jump-starts microbial response to a deep oil spill. Science 330, 208–211 (2010).
Source: Ecology - nature.com



