Morens, D. M. & Fauci, A. S. Emerging Infectious Diseases: Threats to Human Health and Global Stability. Plos Pathog. 9, e1003467 (2013).
Engering, A., Hogerwerf, L. & Slingenbergh, J. Pathogen-host-environment interplay and disease emergence. Emerging Microbes and Infections 2, e5, https://doi.org/10.1038/emi.2013.5 (2013).
Woolhouse, M. E. J., Haydon, D. T. & Antia, R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends in Ecology and Evolution 20, 238–244 (2005).
Lambrechts, L. Dissecting the genetic architecture of host-pathogen specificity. Plos Pathog. 6, 9–10 (2010).
Ribet, D. & Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes and Infection 17, 173–183 (2015).
Pizarro-Cerdá, J. & Cossart, P. Bacterial adhesion and entry into host cells. Cell 124, 715–727 (2006).
Longdon, B., Brockhurst, M. A., Russell, C. A., Welch, J. J. & Jiggins, F. M. The Evolution and Genetics of Virus Host Shifts. Plos Pathog. 10, e1004395, https://doi.org/10.1371/journal.ppat.1004395 (2014).
Vouga, M. & Greub, G. Emerging bacterial pathogens: The past and beyond. Clinical Microbiology and Infection 22, 12–21 (2016).
Pereyre, S., Goret, J. & Bébéar, C. Mycoplasma pneumoniae: Current knowledge on macrolide resistance and treatment. Front. Microbiol. 7, 974, https://doi.org/10.3389/fmicb.2016.00974 (2016).
Maunsell, F. P. & Donovan, G. A. Mycoplasma bovis Infections in Young Calves. Veterinary Clinics of North America – Food Animal Practice 25, 139–177 (2009).
Citti, C. & Blanchard, A. Mycoplasmas and their host: Emerging and re-emerging minimal pathogens. Trends in Microbiology 21, 196–203 (2013).
Rosengarten, R. et al. The Changing Image of Mycoplasmas: From Innocent Bystanders to Emerging and Reemerging Pathogens in Human and Animal Diseases. in Emerging Bacterial Pathogens (eds. Muhldorfer, I. & Shafer, K.) 166–185 (2001).
Sirand-Pugnet, P., Citti, C., Barré, A. & Blanchard, A. Evolution of mollicutes: down a bumpy road with twists and turns. Res. Microbiol. 158, 754–766 (2007).
Rosengarten, R. et al. Host-pathogen interactions in mycoplasma pathogenesis: Virulence and survival strategies of minimalist prokaryotes. Int. J. Med. Microbiol. 290, 15–25 (2000).
Delaney, N. F. et al. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. Plos Genet. 8, e1002511, https://doi.org/10.1371/journal.pgen.1002511 (2012).
Morelli, G. et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. Plos Genet. 6, 1–12 (2010).
Citti, C., Nouvel, L.-X. & Baranowski, E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 5, 1073–1085 (2010).
May, M., Papazisi, L., Gorton, T. S. & Geary, S. J. Identification of fibronectin-binding proteins in Mycoplasma gallisepticum strain R. Infect. Immun. 74, 1777–1785, https://doi.org/10.1128/IAI.74.3.1777-1785.2006 (2006).
Masukagami, Y. et al. The Mycoplasma gallisepticum virulence factor lipoprotein MslA is a novel polynucleotide binding protein. Infect. Immun. 81, 3220–3226 (2013).
Szczepanek, S. M. et al. Identification of lipoprotein MslA as a neoteric virulence factor of Mycoplasma gallisepticum. Infect. Immun. 78, 3475–3483 (2010).
Hudson, P. et al. Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect. Immun. 74, 931–939, https://doi.org/10.1128/IAI.74.2.931-939.2006 (2006).
Winner, F., Rosengarten, R. & Citti, C. In vitro cell invasion of Mycoplasma gallisepticum. Infect. Immun., https://doi.org/10.1128/IAI.68.7.4238-4244.2000 (2000).
Vogl, G. et al. Mycoplasma gallisepticum invades chicken erythrocytes during infection. Infect. Immun. 68, 4238–4244, https://doi.org/10.1128/IAI.00871-07 (2008).
Ley, D. H., Berkhoff, J. E. & McLaren, J. M. Mycoplasma gallisepticum Isolated from House Finches (Carpodacus mexicanus) with Conjunctivitis. Avian Dis. 40, 480–483, https://doi.org/10.2307/1592250 (2006).
Fischer, J. R., Stallknecht, D. E., Luttrell, M. P., Dhondt, A. A. & Converse, K. A. Mycoplasmal Conjunctivitis in Wild Songbirds: The Spread of a New Contagious Disease in a Mobile Host Population. Emerg. Infect. Dis. 3, 69–72, https://doi.org/10.3201/eid0301.970110 (1997).
Levisohn, S. & Kleven, S. H. Avian mycoplasmosis (Mycoplasma gallisepticum). Rev. Sci. Tech. 32, 220–231 (2000).
Ley, D. H., Berkhoff, J. E. & McLaren, J. M. Mycoplasma gallisepticum isolated from house finches (Carpodacus mexicanus) with conjunctivitis. Avian Dis. 40, 480–483 (1996).
Hochachka, W. M. et al. Multiple host transfers, but only one successful lineage in a continent-spanning emergent pathogen. Proc. R. Soc. B Biol. Sci. 280, 20131068, https://doi.org/10.1098/rspb.2013.1068 (2013).
Gumulak-Smith, J. et al. Variations in the surface proteins and restriction enzyme systems of Mycoplasma pulmonis in the respiratory tract of infected rats. Mol. Microbiol. 40, 1037–1044 (2001).
Szczepanek, S. M. et al. Comparative genomic analyses of attenuated strains of Mycoplasma gallisepticum. Infect. Immun. 78, 1760–71 (2010).
Tulman, E. R. et al. Extensive variation in surface lipoprotein gene content and genomic changes associated with virulence during evolution of a novel North American house finch epizootic strain of Mycoplasma gallisepticum. Microbiol. 158, 2073–2088, https://doi.org/10.1099/mic.0.058560-0 (2012).
Papazisi, L., Troy, K. E., Gorton, T. S., Liao, X. & Geary, S. J. Analysis of cytadherence-deficient, GapA-negative Mycoplasma gallisepticum strain R. Infect. Immun. 68, 6643–6649 (2000).
Winner, F. et al. Phenotypic switching in Mycoplasma gallisepticum hemadsorption is governed by a high-frequency, reversible point mutation. Infect. Immun. 71, 1265–1273 (2003).
Indiková, I. et al. Role of the GapA and CrmA Cytadhesins of Mycoplasma gallisepticum in Promoting Virulence and Host Colonization. Infect. Immun. 81, 1618–1624, https://doi.org/10.1128/iai.00112-13 (2013).
Papazisi, L. et al. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain Rlow. Microbiology 149, 2307–2316, https://doi.org/10.1099/mic.0.26427-0 (2003).
Much, P., Winner, F., Stipkovits, L., Rosengarten, R. & Citti, C. Mycoplasma gallisepticum: Influence of cell invasiveness on the outcome of experimental infection in chickens. FEMS Immunol. Med. Microbiol. 15, 181–186, https://doi.org/10.1016/S0928-8244(02)00378-4 (2002).
Ron, M. et al. Mycoplasma gallisepticum in vivo induced antigens expressed during infection in chickens. Vet. Microbiol. 175, 265–274 (2015).
Szczepanek, S. M., Boccaccio, M., Pflaum, K., Liao, X. & Geary, S. J. Hydrogen peroxide production from glycerol metabolism is dispensable for virulence of Mycoplasma gallisepticum in the tracheas of chickens. Infect. Immun. 82, 4915–4920 (2014).
Fürnkranz, U. et al. Factors influencing the cell adhesion and invasion capacity of Mycoplasma gallisepticum. Acta Vet. Scand. 55, 63 (2013).
Hawley, D. M. et al. Parallel Patterns of Increased Virulence in a Recently Emerged Wildlife Pathogen. Plos Biol. 11, e1001570, https://doi.org/10.1371/journal.pbio.1001570 (2013).
Bonneaud, C. et al. Rapid Antagonistic Coevolution in an Emerging Pathogen and Its Vertebrate Host. Curr. Biol. 28, 2978–2983, https://doi.org/10.1016/j.cub.2018.07.003 (2018).
Bonneaud, C. et al. Rapid evolution of disease resistance is accompanied by functional changes in gene expression in a wild bird. Proc. Natl. Acad. Sci. 108, 7866–7871 (2011).
Winner, F., Rosengarten, R. & Citti, C. In vitro cell invasion of Mycoplasma gallisepticum. Infect. Immun. 68, 4238–4244 (2000).
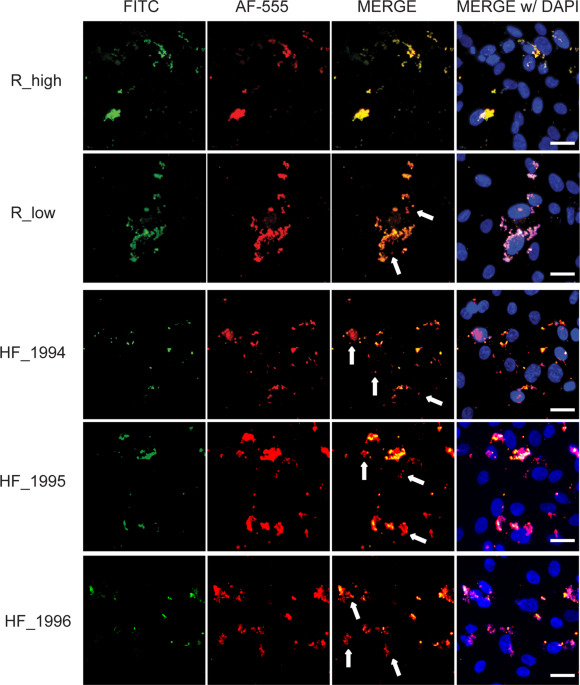
Matyushkina, D. et al. Phase Transition of the Bacterium upon Invasion of a Host Cell as a Mechanism of Adaptation: a Mycoplasma gallisepticum Model. Sci. Rep. 6, 35959 (2016).
Heesemann, J. & Laufs, R. Double immunofluorescence microscopic technique for accurate differentiation of extracellularly and intracellularly located bacteria in cell culture. J. Clin. Microbiol. 22, 168–175 (1985).
Elsinghorst, E. A. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236, 405–420 (1994).
Finlay, B. B. & Cossart, P. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276, 718–725 (1997).
Razin, S. Adherence of pathogenic mycoplasmas to host cells. Biosci. Rep. 19, 367–372 (1999).
Rottem, S. Interaction of mycoplasmas with host cells. Physiol. Rev. 83, 417–432 (2003).
Browning, G. F., Marenda, M. S., Noormohammadi, A. H. & Markham, P. F. The central role of lipoproteins in the pathogenesis of mycoplasmoses. Veterinary Microbiology 153, 44–50 (2011).
Razin, S. & Jacobs, E. Review Article Mycoplasma adhesion. J. Gen. Microbiol. 138, 407–422 (1992).
Goh, M. S., Gorton, T. S., Forsyth, M. H., Troy, K. E. & Geary, S. J. Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA). Microbiology 144, 2971–2978 (1998).
Keeler, C. L., Hnatow, L. L., Whetzel, P. L. & Dohms, J. E. Cloning and characterization of a putative cytadhesin gene (mgc1) from Mycoplasma gallisepticum. Infect. Immun. 64, 1541–1547 (1996).
Yoshida, S., Fujisawa, A., Tsuzaki, Y. & Saitoh, S. Identification and expression of a Mycoplasma gallisepticum surface antigen recognized by a monoclonal antibody capable of inhibiting both growth and metabolism. Infect. Immun. 68, 3186–3192 (2000).
Tarshis, M., Yavlovich, A., Katzenell, A., Ginsburg, I. & Rottem, S. Intracellular Location and Survival of Mycoplasma penetrans Within HeLa Cells. Curr. Microbiol. 49, 136–140 (2004).
Razin, S., Yogev, D. & Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–156 (1998).
Cecchini, K. R., Gorton, T. S. & Geary, S. J. Transcriptional responses of Mycoplasma gallisepticum strain R in association with eukaryotic cells. J. Bacteriol. 189, 5803–5807, https://doi.org/10.1128/JB.00667-07 (2007).
Hybiske, K. & Stephens, R. S. Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 6, 99–110 (2008).
Hybiske, K. & Stephens, R. S. Mechanisms of host cell exit by the intracellular bacterium. Chlamydia. Proc. Natl. Acad. Sci. USA 104, 11430–11435 (2007).
Lai, X. H., Golovliov, I. & Sjöstedt, A. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69, 4691–4694 (2001).
Yoshida, S. et al. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science 314, 985–989 (2006).
Robbins, J. R. et al. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. J. Cell Biol. 146, 1333–1349 (1999).
Hopfe, M., Deenen, R., Degrandi, D., Köhrer, K. & Henrich, B. Host cell responses to persistent mycoplasmas – different stages in infection of HeLa cells with Mycoplasma hominis. Plos one 8, e54219 (2013).
Bischofberger, M., Iacovache, I. & van der Goot, F. G. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe 12, 266–275 (2012).
Zahrt, T. C. Molecular mechanisms regulating persistent Mycobacterium tuberculosis infection. Microbes Infect. 5, 159–167 (2003).
Shames, S. R. et al. The pathogenic E. coli type III effector EspZ interacts with host CD98 and facilitates host cell prosurvival signalling. Cell. Microbiol. 12, 1322–1339 (2010).
Roxas, J. L. et al. The enteropathogenic Escherichia coli-secreted protein EspZ inhibits host cell apoptosis. Infect. Immun. 80, 3850–3857 (2012).
Koziel, J. et al. The role of Mcl-1 in S. aureus-induced cytoprotection of infected macrophages. Mediators Inflamm. 2013, 427021 (2013).
Logunov, D. Y. et al. Mycoplasma infection suppresses p53, activates NF-kappaB and cooperates with oncogenic Ras in rodent fibroblast transformation. Oncogene 27, 4521–4531 (2008).
Feng, S. H., Tsai, S., Rodriguez, J. & Lo, S. C. Mycoplasmal infections prevent apoptosis and induce malignant transformation of interleukin-3-dependent 32D hematopoietic cells. Mol. Cell. Biol. 19, 7995–8002 (1999).
Xu, J. et al. Mycoplasma gallisepticum MGA_0676 is a membrane-associated cytotoxic nuclease with a staphylococcal nuclease region essential for nuclear translocation and apoptosis induction in chicken cells. Appl. Microbiol. Biotechnol. 99, 1859–1871 (2014).
Gerlic, M., Horowitz, J., Farkash, S. & Horowitz, S. The inhibitory effect of Mycoplasma fermentans on tumour necrosis factor (TNF)-alpha-induced apoptosis resides in the membrane lipoproteins. Cell. Microbiol. 9, 142–153 (2007).
Bischof, D. F., Janis, C., Vilei, E. M., Bertoni, G. & Frey, J. Cytotoxicity of Mycoplasma mycoides subsp. mycoides small colony type to bovine epithelial cells. Infect. Immun. 76, 263–269, https://doi.org/10.1128/IAI.00938-07 (2008).
Rottem, S. Invasion of Mycoplasmas and Fusion with Host Cells. Molecular Biology and Pathogenicity of Mycoplasmas (Kluwer Academic, 2002).
R Development Core Team, R. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing 1 (2011).
Wickham, H. Ggplot2. Elegant Graphics for Data Analysis, https://doi.org/10.1007/978-0-387-98141-3 (2009).
Source: Ecology - nature.com



