On the Namibian continental shelf, Foraminifera live below the seafloor down to ca. 28 cmbsf in an anoxic environment that is extremely high in sulfide [26]. The dominance of Bolivina throughout the core and our detection of their 18S rRNA, even into the anoxic depths, is consistent with the known affinity of Bolivina for oxygen-depleted habitats [27], including the studied region as it was observed previously in sulfidic sediments at multiple coring locations on the Namibian shelf [26]. The “trophic oxygen model” predicts that the dynamic nature of microhabitats allows Foraminifera to migrate up and down in the sediment with the prevailing redox conditions [28]. Hence, since we sampled during the southern Winter when bottom water oxygen levels in the Namibian OMZ are higher [29, 30], it is possible that the penetration depth of the Foraminifera extended relatively deep because of the higher oxygen concentration at the sediment surface.
Although the diversity of Foraminifera is well constrained by morphological studies, the group is not yet well represented in transcriptomic and genomic databases. The recently large transcriptome sequencing effort of microbial eukaryotes helped to alleviate this problem [14], since it included several Foraminifera that we could add to our database. Nevertheless, because of the relatively low number of sequenced genomes and transcriptomes from Foraminifera (compared with bacteria for example), our metatranscriptome approach cannot distinguish between ORFs derived from different Foraminifera species. The ORFs assigned to Foraminifera here thus serves as a “group averaging”, but should correspond to genetically similar populations since the de novo assemblies that are used to build the contigs from the RNA-seq data are based on genetic similarity (see “Methods”). Furthermore, our metatranscriptomes contained the complete 18S rRNA sequence (Fig. 3) from the most abundant taxa, i.e., Bolivina sp. and Stainforthia sp. (Figs. 1, 3, and S2) and thus we are confident that the ORFs assigned as Foraminifera are derived primarily from these cytoplasm-containing Foraminifera tests that we could enumerate in the core (Fig. 1). Despite the presence of two morphological different Bolivina species in the core, we could not find signs for the active expression of the 18S rRNA in the second species. This indicates that most of the identified foraminiferan metatranscriptomic expression likely comes from one of the Bolivina species in addition to Stainforthia sp.
The composition of the community is important when using relative abundances for normalization to compare between samples, since the comparison can be biased if certain groups are present at high abundance in some samples, and missing in others. In our sampled core however, all the same major protist groups were present at all depths, albeit in different proportions (Fig. 2c), indicating that differences in relative abundance of groups can be compared between depths. Thus, the differences in relative levels of gene expression of Foraminifera between depths cannot be explained solely by a different community composition at those depths. Similarly, in this same core the composition of the bacterial and archaeal community also does not change drastically with sediment depth [31]. A higher number of unique ORFs would be expected to increase with a higher proportion of Foraminifera as this is associated with increased sequencing depth leading to the recovery of more of the less expressed ORFs. However, the concentration of Foraminifera cells is six times lower in the deepest anoxic samples (Fig. 1), but at this depth the Foraminifera have eight- to tenfold higher relative levels of gene expression compared with those at the surface (Fig. 2). An eight- to tenfold increase in gene expression, from a community with a sixfold lower concentration of cytoplasm-containing cells, is a strong indication for increased transcriptional activity per Foraminifera cell. There was a relatively higher percentage of Foraminifera transcripts involved in energy production (KOG:C) in the surface sample, compared with the deeper anoxic samples (Fig. 2d). We speculate that this could be due to the availability of O2 at the sediment seawater interface, which can fuel aerobic respiration and increased energy metabolism.
Foraminifera are predators, and are thought to act primarily as heterotrophs utilizing ingested prey cells as carbon sources for growth [32]. Our gene expression analysis provides insights into the possible mechanisms of prey acquisition, and the metabolic processing of the ingested material. The expression of ORFs encoding Rho proteins by Foraminifera indicate an active induction of phagocytosis, since Rho proteins function in actin dynamics during phagocytosis [20, 21]. Myosin motor proteins are recruited to the cell membrane during phagocytosis in order to envelope and capture prey particles [33], and the prey then enter the phagocytosing cell as a food vacuole [25]. Food vacuoles are then transported into the cell via dynein along microtubules, where the contents are digested under acidic conditions via the activity of vacuolar-type H+ ATPases [25] (Fig. 4c). Such proton pumping ATPases are responsible for lysing digested prey cells inside food vacuoles after phagocytosis, where the acidified lysosomal vesicles are loaded with digestive enzymes [25]. The metatranscriptome data indicate that under anoxic conditions, the Foraminifera metabolize the hydrolyzed organics for ATP production via fermentation and fumarate reduction, and dissimilatory nitrite reduction (Fig. 4c). Because cells are mostly protein, anaerobic fermentation of ingested prey cells by Foraminifera may include amino acid fermentations. By weight, exponentially growing cells are made of roughly 50–60% protein, 20% RNA, 10% lipids, 3% DNA, 10–20% sugars as cell wall constituents, and some metabolites [34]. Amino acid fermentations provide roughly one net ATP per amino acid fermented [25].
The fumarate reduction during anaerobic energy metabolism in eukaryotes is usually associated with rhodoquinone (RQ) as an electron carrier, and RQ generally replaces ubiquinone as an electron carrier in the electron transport chain after the switch from aerobic metabolism to anaerobic metabolism [35]. The switch to RQ synthesis during anaerobic metabolism is controlled by the polyprenyltransferase COQ-2 [35], but we could not find any expressed ORFs in our metatranscriptomes with significant similarity to this gene. Future controlled experiments involving the switch from anaerobic conditions could test whether Foraminifera indeed use RQ as an electron carrier during anaerobic energy metabolism.
In addition to hydrolyzed organics from ingested prey, the transcriptomes suggest that CME is another mechanism by which Foraminifera could utilize both high- and low-molecular weight dissolved organic matter (dissolved in the pore water of the sediments) under anoxic conditions. Experiments using 13C-labeled diatom prey showed that under anoxic conditions the benthic foram Ammonia tepida reduced the number of phagocytosed diatom cells, and the ingested cells were apparently not digested inside vacuoles but remained intact after 4 weeks [36]. If a decreased utilization of ingested prey for energy production is a general feature of anaerobic Foraminifera, it is possible that organic matter obtained via CME (Fig. 4b, c) becomes a relatively more important carbon source as opposed to ingested prey cells.
Eukaryotic fermentations can produce a variety of end products, and our data indicate the possibility for Foraminifera to produce ethanol, acetate, and succinate (Fig. 4c). Under conditions of prolonged anaerobiosis, propionate is preferentially formed as opposed to succinate in anaerobic mitochondria, whereby one additional ATP and one CO2 are formed from d-methylmalonyl-CoA via propionyl-CoA carboxylase [25, 37]. We detected expression of a Foraminifera ORF with similarity to propionyl-CoA carboxylase at 28 cmbsf indicating that prolonged anoxic conditions in the sulfidic sediments at 28 cm stimulated production of propionate in Foraminifera mitochondria.
A key intermediate in the anaerobic energy metabolism of most eukaryotes is malate [12, 37]. During anaerobic respiration in many eukaryotes malate is converted to fumarate via the enzyme fumarase running in reverse, and the resulting fumarate then can be used as the terminal electron acceptor [12, 37]. This fumarate reduction is coupled to an anaerobic electron transport chain in which electrons are transferred from NADH to fumarate via a specialized complex I and a mitochondrial membrane associated fumarate reductase [12, 37]. This physiology is typical of anaerobic mitochondria, that are widely distributed amongst eukaryotes including Foraminifera, Bivalvia, Polychaeta, Platyhelminthes, Nematoda, Euglenida, and Ciliophora [12].
The metatranscriptomes furthermore indicated that under anoxic conditions, Foraminifera utilize creatine kinase and phosphocreatine to maintain cellular energy homeostasis (Fig. 4c). In many eukaryotic cells, creatine kinase acts as a mechanism for maintaining balance between ATP consuming and producing processes [38]. Our data indicate that this also occurs in anaerobic Foraminifera. In human cells, creatine kinase acts as an ATP regenerator, and the phosphocreatine pool is used as a temporal energy buffer to maintain ATP/ADP ratios inside the cell [38]. By acting as an energy shuttle between ATP providing and consuming processes, phosphocreatine acts as a phosphogen to maintain the concentration of the high-energy phosphate pool inside the cell. This facilitates more energetically costly cellular activities under anoxic conditions, such as phagocytosis, by maintaining the spatial “energy circuit” [39]. For example, creatine kinase contributes to the build-up of a large intracellular pool of phosphocreatine that represents an efficient temporal energy buffer and prevents a rapid fall in global ATP concentrations [38]. This likely helps to couple the energy producing and energy consuming processes inside of Foraminifera cells during anaerobic metabolism. An increased utilization of creatine kinase under anoxic conditions is supported by the 10-day incubation, whereby expression of Foraminifera ORFs with highest similarity to creatine kinases, as well as enzymes involving in mitochondrial energy production, increased with the onset of anoxic conditions (Fig. 6).
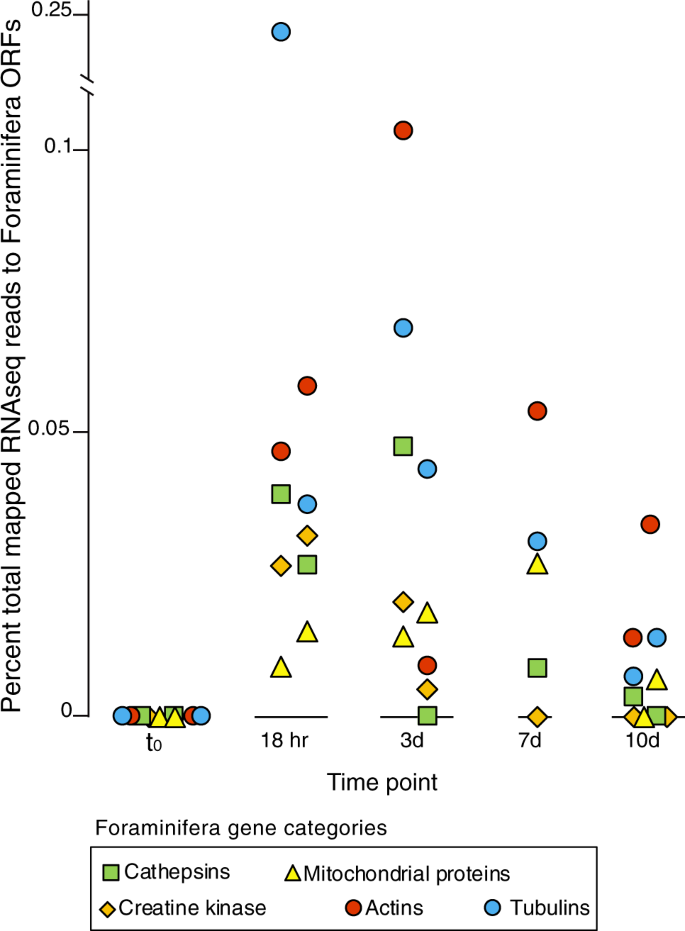
The five gene categories are shown based on KOG annotations. The category “mitochondrial proteins” are those KOG annotations that have the word “mitochondria”, “mitochondrion”, or “mitochondrial”, in the KOG description. The replicates are shown for each time point are displayed as two individual points, and represent the fractional abundance of all reads mapping to ORFs with a given annotation.
Biogeochemical studies indicate that foraminiferans are capable of performing denitrification, that is, the conversion of NO3− to N2 [9]. The enzymes behind the foraminiferal denitrification pathway in the genus Globobulimina appear to be acquired relatively early in Foraminifera evolution [10], and it was indicated that the foraminifera themselves, not associated prokaryotes, are performing the denitrification reaction [40, 41]. The sequestration of nitrate by Foraminifera is highly suggestive that the protists themselves, and not associated symbionts, are performing nitrate respiration [40, 41].
Consistent with this prior evidence, we found the genes of the denitrification pathway in Foraminifera to be expressed (Fig. 4), including a putative assimilatory nitrate reductase (Nr). This may function as a sulfite oxidase or dissimilatory nitrate reductase [10, 42]. We interpret the Nr genes to be involved in dissimilatory nitrate reduction with caution and refer to them as “putative nitrate reductases” since it is possible that the Nr genes function solely for nitrate assimilation in Foraminifera [10]. In any case, our data show that these Nr genes are transcribed during anaerobic metabolism in benthic Foraminifera.
The expression of nitrate transporters [10] from Foraminifera at 28 cmbsf (Fig. 4a) seems contradictory to the geochemical conditions, since nitrate and nitrite were both below detection at this depth in the core (Fig. 1). However, this can be explained by the fact that many benthic Foraminifera can store nitrate in vacuoles under anoxic conditions and use the stored nitrate and nitrite as terminal electron acceptors for anaerobic respiration [9, 40, 41]. Thus, the expression of the nitrate transporter genes seen here could be responsible for transporting nitrate out of the vacuole (and regulating the cytosolic concentration of nitrate), and into the mitochondrion, as has been proposed previously for denitrifying Foraminifera based on genome data [10]. The expression of the NirK and Nor genes indicate that the Foraminifera were actively performing two key steps of denitrification—nitrite and nitric oxide reduction (Fig. 4c). Some Bolivina and Stainforthia and species lack a nitrous oxide reductase and reduce nitrate only to N2O [40, 41, 43], and we did not detect any expression of NosZ indicating that the denitrifying Bolivina and Stainforthia species in our samples were also likely reducing nitrite to nitric oxide, that is then reduced to N2O via Nor (Fig. 4c). The lack of expression of the NosZ gene raises the possibility that the denitrifying Foraminifera in Namibian sediments are a source of N2O, an important greenhouse gas [44].
The high levels of sulfide in the Namibian sediments should be toxic to eukaryotic life and thus similar to other eukaryotes it would be expected that mechanisms exist to help Foraminifera cope with these conditions. For example, eukaryotic sulfide quinone reductase (SQR) [45] and the sulfide resistant alternative oxidase (AOX) are used by some animals and protozoa to modify their metabolism under anoxic conditions and survive the sulfidic conditions [12]. The KOG database does not contain the SQR, nor AOX genes. After adding all available SQR and AOX genes available in the NCBI Protein database to the KOG database, DIAMOND [46] searches detected one ORF at 12 cmbsf with highest similarity to the Foraminifera Ammonia that had significant similarity (amino acid similarity: 46%, alignment length: 259 amino acids, e-value: 10−27) to AOX from the Oomycete Pythium aphanidermatum (Genbank Accession: CAE11918.1). This indicates that some Foraminifera use AOX to perform aerobic fermentation, similar to parasitic trypanosomes, whereby O2 is used as the terminal electron acceptor to reoxidize ubiquinol for pyrimidine biosynthesis—as opposed to mitochondrial ATP synthesis [12]. AOX requires O2, which we speculate could be available in limited, ephemeral concentrations from the bioirrigating worms that were observed in the core.
The large increase in Foraminifera gene expression upon the onset of anoxic conditions in the incubation (Fig. 5b–d) provides experimental support for the observation of increasing Foraminifera gene expression with increasing depths and sulfidic conditions in the core (Fig. 2c). The higher number of ORFs expressed by Foraminifera after the onset of anoxia (Fig. 5c) indicates that some Foraminifera increased the number of expressed genes, rather than the increase being due to the dying off of other eukaryotes causing an increased relative abundance of Foraminifera transcripts. The increased number of ORFs expressed by Foraminifera could be primarily attributed to those involved in the cytoskeleton, translation, posttranslational modification, and intracellular trafficking (Fig. 5c). This indicates that many Foraminifera were modifying their physiology, increasing translation and protein synthesis in response to anoxic conditions. An increased expression of Foraminifera genes involved in production and modification of the cytoskeleton also suggests that anoxia increased cellular activity [47], rather than causing an increased expression of stress related genes due to unfavorable conditions like the accumulation of hydrogen sulfide. Indeed, no SQR was expressed by Foraminifera during the incubations, which are used by many eukaryotes to cope with sulfidic conditions [12].
Our findings demonstrate that activity of benthic Foraminifera in these sulfidic Namibian sediments is stimulated by anoxic conditions, similar to the findings of a metabolic preference of nitrate over oxygen as an electron acceptor in the Peruvian oxygen minimum zone [11]. The peak stimulation of Foraminifera gene expression after 18 h at the onset of anoxic conditions might indicate the utilization of fumarate, nitrate, and or nitrite by anaerobic denitrifying foraminifera as terminal electron acceptors. This indicates that the Bolivina and Stainforthia species in the Namibian sediments are anaerobes that prefer anoxic conditions, as this clearly stimulated their activity compared with aerobic conditions.
Source: Ecology - nature.com



